Retinal Ganglion Cell Diversity and Subtype Specification from Human Pluripotent Stem Cells
- PMID: 29576537
- PMCID: PMC5998302
- DOI: 10.1016/j.stemcr.2018.02.010
Retinal Ganglion Cell Diversity and Subtype Specification from Human Pluripotent Stem Cells
Abstract
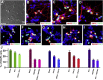
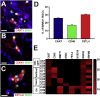
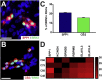
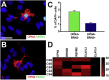
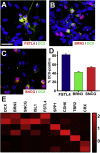
Retinal ganglion cells (RGCs) are the projection neurons of the retina and transmit visual information to postsynaptic targets in the brain. While this function is shared among nearly all RGCs, this class of cell is remarkably diverse, comprised of multiple subtypes. Previous efforts have identified numerous RGC subtypes in animal models, but less attention has been paid to human RGCs. Thus, efforts of this study examined the diversity of RGCs differentiated from human pluripotent stem cells (hPSCs) and characterized defined subtypes through the expression of subtype-specific markers. Further investigation of these subtypes was achieved using single-cell transcriptomics, confirming the combinatorial expression of molecular markers associated with these subtypes, and also provided insight into more subtype-specific markers. Thus, the results of this study describe the derivation of RGC subtypes from hPSCs and will support the future exploration of phenotypic and functional diversity within human RGCs.
Keywords: RGC subtype; RNA-seq; alpha RGC; direction selective RGC; iPSC; ipRGC; retina; retinal ganglion cell; stem cell.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Berson D.M. Retinal ganglion cell types and their central projections. In: Masland R.H., Albright T.D., editors. Vol. 1. Elsevier; 2008. pp. 491–519. (The Senses: A Comprehensive Reference).
-
- Chew K.S., Renna J.M., McNeill D.S., Fernandez D.C., Keenan W.T., Thomsen M.B., Ecker J.L., Loevinsohn G.S., VanDunk C., Vicarel D.C. A subset of ipRGCs regulates both maturation of the circadian clock and segregation of retinogeniculate projections in mice. Elife. 2017;6 https://doi.org/10.7554/eLife.22861 - DOI - PMC - PubMed
-
- Croner L.J., Kaplan E. Receptive fields of P and M ganglion cells across the primate retina. Vision Res. 1995;35:7–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

