Human iPSC-Derived Endothelial Cells and Microengineered Organ-Chip Enhance Neuronal Development
- PMID: 29576540
- PMCID: PMC5998748
- DOI: 10.1016/j.stemcr.2018.02.012
Human iPSC-Derived Endothelial Cells and Microengineered Organ-Chip Enhance Neuronal Development
Abstract
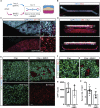
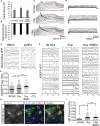
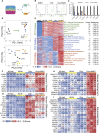
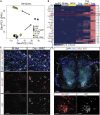
Human stem cell-derived models of development and neurodegenerative diseases are challenged by cellular immaturity in vitro. Microengineered organ-on-chip (or Organ-Chip) systems are designed to emulate microvolume cytoarchitecture and enable co-culture of distinct cell types. Brain microvascular endothelial cells (BMECs) share common signaling pathways with neurons early in development, but their contribution to human neuronal maturation is largely unknown. To study this interaction and influence of microculture, we derived both spinal motor neurons and BMECs from human induced pluripotent stem cells and observed increased calcium transient function and Chip-specific gene expression in Organ-Chips compared with 96-well plates. Seeding BMECs in the Organ-Chip led to vascular-neural interaction and specific gene activation that further enhanced neuronal function and in vivo-like signatures. The results show that the vascular system has specific maturation effects on spinal cord neural tissue, and the use of Organ-Chips can move stem cell models closer to an in vivo condition.
Keywords: amyotrophic lateral sclerosis; brain microvascular endothelial cells; disease modeling; iPSC; microfluidic device; microphysiological system; organ-on-chip; spinal cord; spinal motor neurons; vasculature.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. - PubMed
-
- Batra R., Hutt K., Vu A., Rabin S.J., Baughn M.W., Libby R.T., Hoon S., Ravits J., Yeo G.W. Gene expression signatures of sporadic ALS motor neuron populations. bioRxiv. 2016
-
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

