Developing novel methods to image and visualize 3D genomes
- PMID: 29577183
- PMCID: PMC6133007
- DOI: 10.1007/s10565-018-9427-z
Developing novel methods to image and visualize 3D genomes
Abstract
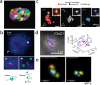
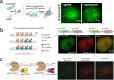
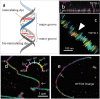
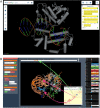
To investigate three-dimensional (3D) genome organization in prokaryotic and eukaryotic cells, three main strategies are employed, namely nuclear proximity ligation-based methods, imaging tools (such as fluorescence in situ hybridization (FISH) and its derivatives), and computational/visualization methods. Proximity ligation-based methods are based on digestion and re-ligation of physically proximal cross-linked chromatin fragments accompanied by massively parallel DNA sequencing to measure the relative spatial proximity between genomic loci. Imaging tools enable direct visualization and quantification of spatial distances between genomic loci, and advanced implementation of (super-resolution) microscopy helps to significantly improve the resolution of images. Computational methods are used to map global 3D genome structures at various scales driven by experimental data, and visualization methods are used to visualize genome 3D structures in virtual 3D space-based on algorithms. In this review, we focus on the introduction of novel imaging and visualization methods to study 3D genomes. First, we introduce the progress made recently in 3D genome imaging in both fixed cell and live cells based on long-probe labeling, short-probe labeling, RNA FISH, and the CRISPR system. As the fluorescence-capturing capability of a particular microscope is very important for the sensitivity of bioimaging experiments, we also introduce two novel super-resolution microscopy methods, SDOM and low-power super-resolution STED, which have potential for time-lapse super-resolution live-cell imaging of chromatin. Finally, we review some software tools developed recently to visualize proximity ligation-based data. The imaging and visualization methods are complementary to each other, and all three strategies are not mutually exclusive. These methods provide powerful tools to explore the mechanisms of gene regulation and transcription in cell nuclei.
Keywords: 3D genomes; Chromatins; FISH method.
Figures
Similar articles
-
ChromoTrace: Computational reconstruction of 3D chromosome configurations for super-resolution microscopy.PLoS Comput Biol. 2018 Mar 9;14(3):e1006002. doi: 10.1371/journal.pcbi.1006002. eCollection 2018 Mar. PLoS Comput Biol. 2018. PMID: 29522506 Free PMC article.
-
3D genome and its disorganization in diseases.Cell Biol Toxicol. 2018 Oct;34(5):351-365. doi: 10.1007/s10565-018-9430-4. Epub 2018 May 23. Cell Biol Toxicol. 2018. PMID: 29796744 Review.
-
Three-dimensional organization and dynamics of the genome.Cell Biol Toxicol. 2018 Oct;34(5):381-404. doi: 10.1007/s10565-018-9428-y. Epub 2018 Mar 22. Cell Biol Toxicol. 2018. PMID: 29568981 Free PMC article. Review.
-
3D Multicolor DNA FISH Tool to Study Nuclear Architecture in Human Primary Cells.J Vis Exp. 2020 Jan 25;(155). doi: 10.3791/60712. J Vis Exp. 2020. PMID: 32065142
-
Image analysis workflows to reveal the spatial organization of cell nuclei and chromosomes.Nucleus. 2022 Dec;13(1):277-299. doi: 10.1080/19491034.2022.2144013. Nucleus. 2022. PMID: 36447428 Free PMC article.
Cited by
-
Recent Progress in Small Spirocyclic, Xanthene-Based Fluorescent Probes.Molecules. 2020 Dec 16;25(24):5964. doi: 10.3390/molecules25245964. Molecules. 2020. PMID: 33339370 Free PMC article. Review.
-
Investigating epithelial-to-mesenchymal transition with integrated computational and experimental approaches.Phys Biol. 2019 Mar 7;16(3):031001. doi: 10.1088/1478-3975/ab0032. Phys Biol. 2019. PMID: 30665206 Free PMC article. Review.
-
Association of mutation and low expression of the CTCF gene with breast cancer progression.Saudi Pharm J. 2020 May;28(5):607-614. doi: 10.1016/j.jsps.2020.03.013. Epub 2020 Apr 2. Saudi Pharm J. 2020. PMID: 32435142 Free PMC article.
-
Background-suppressed live visualization of genomic loci with an improved CRISPR system based on a split fluorophore.Genome Res. 2020 Sep;30(9):1306-1316. doi: 10.1101/gr.260018.119. Epub 2020 Sep 4. Genome Res. 2020. PMID: 32887690 Free PMC article.
-
Advances of super-resolution fluorescence polarization microscopy and its applications in life sciences.Comput Struct Biotechnol J. 2020 Jun 26;18:2209-2216. doi: 10.1016/j.csbj.2020.06.038. eCollection 2020. Comput Struct Biotechnol J. 2020. PMID: 32952935 Free PMC article. Review.
References
-
- Beliveau BJ, Joyce EF, Apostolopoulos N, Yilmaz F, Fonseka CY, McCole RB, Chang YM, Li JB, Senaratne TN, Williams BR, et al. Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. P Natl Acad Sci USA. 2012;109:21301–21306. doi: 10.1073/pnas.1213818110. - DOI - PMC - PubMed
-
- Beliveau BJ, Boettiger AN, Avendano MS, Jungmann R, McCole RB, Joyce EF, Kim-Kiselak C, Bantignies F, Fonseka CY, Erceg J, et al. Single-molecule super-resolution imaging of chromosomes and in situ haplotype visualization using Oligopaint FISH probes. Nat Commun. 2015;6:7147. doi: 10.1038/ncomms8147. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

