Multi-Invasion-Induced Rearrangements as a Pathway for Physiological and Pathological Recombination
- PMID: 29578583
- PMCID: PMC6072258
- DOI: 10.1002/bies.201700249
Multi-Invasion-Induced Rearrangements as a Pathway for Physiological and Pathological Recombination
Abstract
Cells mitigate the detrimental consequences of DNA damage on genome stability by attempting high fidelity repair. Homologous recombination templates DNA double-strand break (DSB) repair on an identical or near identical donor sequence in a process that can in principle access the entire genome. Other physiological processes, such as homolog recognition and pairing during meiosis, also harness the HR machinery using programmed DSBs to physically link homologs and generate crossovers. A consequence of the homology search process by a long nucleoprotein filament is the formation of multi-invasions (MI), a joint molecule in which the damaged ssDNA has invaded more than one donor molecule. Processing of MI joint molecules can compromise the integrity of both donor sites and lead to their rearrangement. Here, two mechanisms for the generation of rearrangements as a pathological consequence of MI processing are detailed and the potential relevance for non-allelic homologous recombination discussed. Finally, it is proposed that MI-induced crossover formation may be a feature of physiological recombination.
Keywords: copy number variation; genomic instability; homologous recombination; non-allelic homologous recombination; repeated sequence; structural variant; translocation.
© 2018 WILEY Periodicals, Inc.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures

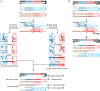

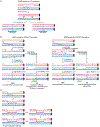

References
-
- Bell JC, Kowalczykowski SC, Annu. Rev. Biochem 2016, 85, 193. - PubMed

