Identifying Small-Molecule Binding Sites for Epigenetic Proteins at Domain-Domain Interfaces
- PMID: 29578648
- PMCID: PMC6001751
- DOI: 10.1002/cmdc.201800030
Identifying Small-Molecule Binding Sites for Epigenetic Proteins at Domain-Domain Interfaces
Abstract
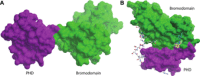
Epigenetics is a rapidly growing field in drug discovery. Of particular interest is the role of post-translational modifications to histones and the proteins that read, write, and erase such modifications. The development of inhibitors for reader domains has focused on single domains. One of the major difficulties of designing inhibitors for reader domains is that, with the notable exception of bromodomains, they tend not to possess a well-enclosed binding site amenable to small-molecule inhibition. As many of the proteins in epigenetic regulation have multiple domains, there are opportunities for designing inhibitors that bind at a domain-domain interface which provide a more suitable interaction pocket. Examination of X-ray structures of multiple domains involved in recognising and modifying post-translational histone marks using the SiteMap algorithm identified potential binding sites at domain-domain interfaces. For the tandem plant homeodomain-bromodomain of SP100C, a potential inter-domain site identified computationally was validated experimentally by the discovery of ligands by X-ray crystallographic fragment screening.
Keywords: X-ray fragment screening; bromodomains; epigenetics; histones; tudor domains.
© 2018 The Authors. Published by Wiley-VCH Verlag GmbH & Co. KGaA.
Figures





References
-
- Jenuwein T., Allis C. D., Science 2001, 293, 1074–1080. - PubMed
-
- InterPro: Protein Sequence Analysis & Classification, http://www.ebi.ac.uk/interpro/.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

