Quantitative single-cell transcriptomics
- PMID: 29579145
- PMCID: PMC6063296
- DOI: 10.1093/bfgp/ely009
Quantitative single-cell transcriptomics
Abstract
Single-cell RNA sequencing (scRNA-seq) is currently transforming our understanding of biology, as it is a powerful tool to resolve cellular heterogeneity and molecular networks. Over 50 protocols have been developed in recent years and also data processing and analyzes tools are evolving fast. Here, we review the basic principles underlying the different experimental protocols and how to benchmark them. We also review and compare the essential methods to process scRNA-seq data from mapping, filtering, normalization and batch corrections to basic differential expression analysis. We hope that this helps to choose appropriate experimental and computational methods for the research question at hand.
Figures
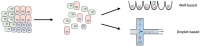
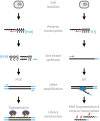

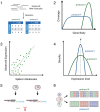

References
-
- Reinius B, Sandberg R.. Random monoallelic expression of autosomal genes: stochastic transcription and allele-level regulation. Nat Rev Genet 2015;16:653–64. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

