ViFi: accurate detection of viral integration and mRNA fusion reveals indiscriminate and unregulated transcription in proximal genomic regions in cervical cancer
- PMID: 29579309
- PMCID: PMC6283451
- DOI: 10.1093/nar/gky180
ViFi: accurate detection of viral integration and mRNA fusion reveals indiscriminate and unregulated transcription in proximal genomic regions in cervical cancer
Abstract
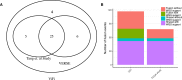
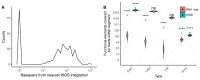
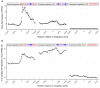
The integration of viral sequences into the host genome is an important driver of tumorigenesis in many viral mediated cancers, notably cervical cancer and hepatocellular carcinoma. We present ViFi, a computational method that combines phylogenetic methods with reference-based read mapping to detect viral integrations. In contrast with read-based reference mapping approaches, ViFi is faster, and shows high precision and sensitivity on both simulated and biological data, even when the integrated virus is a novel strain or highly mutated. We applied ViFi to matched genomic and mRNA data from 68 cervical cancer samples from TCGA and found high concordance between the two. Surprisingly, viral integration resulted in a dramatic transcriptional upregulation in all proximal elements, including LINEs and LTRs that are not normally transcribed. This upregulation is highly correlated with the presence of a viral gene fused with a downstream human element. Moreover, genomic rearrangements suggest the formation of apparent circular extrachromosomal (ecDNA) human-viral structures. Our results suggest the presence of apparent small circular fusion viral/human ecDNA, which correlates with indiscriminate and unregulated expression of proximal genomic elements, potentially contributing to the pathogenesis of HPV-associated cervical cancers. ViFi is available at https://github.com/namphuon/ViFi.
Figures





References
-
- Plummer M., de Martel C., Vignat J., Ferlay J., Bray F., Franceschi S.. Global burden of cancers attributable to infections in 2012: a synthetic analysis. Lancet Glob. Health. 2016; 4:e609–e616. - PubMed
-
- Duensing S., Münger K.. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002; 62:7075–7082. - PubMed
-
- Zhang T., Zhang J., You X., Liu Q., Du Y., Gao Y., Shan C., Kong G., Wang Y., Yang X. et al. . Hepatitis B virus X protein (HBx) modulates oncogene YAP via CREB to promote growth of hepatoma cells. Hepatology. 2012; 56:2051–2059. - PubMed
-
- Carrillo-Infante C., Abbadessa G., Bagella L., Giordano A.. Viral infections as a cause of cancer (review). Int. J. Oncol. 2007; 30:1521–1528. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

