Satellite DNA evolution: old ideas, new approaches
- PMID: 29579574
- PMCID: PMC5975084
- DOI: 10.1016/j.gde.2018.03.003
Satellite DNA evolution: old ideas, new approaches
Abstract
A substantial portion of the genomes of most multicellular eukaryotes consists of large arrays of tandemly repeated sequence, collectively called satellite DNA. The processes generating and maintaining different satellite DNA abundances across lineages are important to understand as satellites have been linked to chromosome mis-segregation, disease phenotypes, and reproductive isolation between species. While much theory has been developed to describe satellite evolution, empirical tests of these models have fallen short because of the challenges in assessing satellite repeat regions of the genome. Advances in computational tools and sequencing technologies now enable identification and quantification of satellite sequences genome-wide. Here, we describe some of these tools and how their applications are furthering our knowledge of satellite evolution and function.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
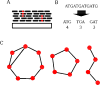
References
-
- John B, Miklos GL. Functional aspects of satellite DNA and heterochromatin. Int Rev Cytol. 1979;58:1–114. - PubMed
-
- Choo KH, Vissel B, Earle E. Evolution of α-satellite DNA on human acrocentric chromosomes. Genomics. 5:332–344. 1989/8. - PubMed
-
- Camacho JPM, Ruiz-Ruano FJ, Martín-Blázquez R, López-León MD, Cabrero J, Lorite P, Cabral-de-Mello DC, Bakkali M. A step to the gigantic genome of the desert locust: chromosome sizes and repeated DNAs. Chromosoma. 2015;124:263–275. - PubMed
-
- Kit S. Equilibrium sedimentation in density gradients of DNA preparations from animal tissues. J Mol Biol. 1961;3:711–IN2. - PubMed
-
- Khost DE, Eickbush DG, Larracuente AM. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res. 2017;27:709–721. The authors employ PacBio sequencing of Drosophila melanogaster to achieve detailed assemblies of the major autosomal complex satellite DNA loci, particularly Responder and 1.688 gm/cm3. They show that PacBio assembly with error correction can significantly extend contigs into repetitive pericentromeric regions. They urge careful examination of assemblies across multiple parameters, using orthogonal molecular and genomic evidence to validate assemblies to ensure accuracy. Their data support models of TE gain at the ends of arrays and are consistent with the homogenizing effect of concerted evolution. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

