New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm
- PMID: 29580211
- PMCID: PMC5870212
- DOI: 10.1186/s12864-018-4602-4
New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm
Abstract
Background: Exploring whether and how mutation of silk protein contributes to subsequent re-allocation of nitrogen, and impacts on the timing of silk gland degradation, is important to understand silk gland biology. Rapid development and wide application of genome editing approach in the silkworm provide us an opportunity to address these issues.
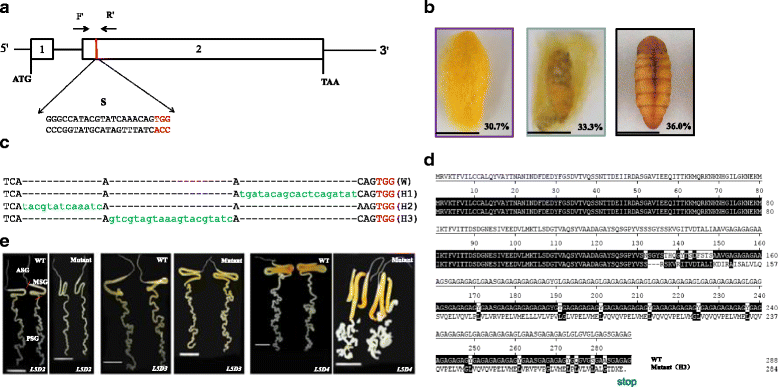
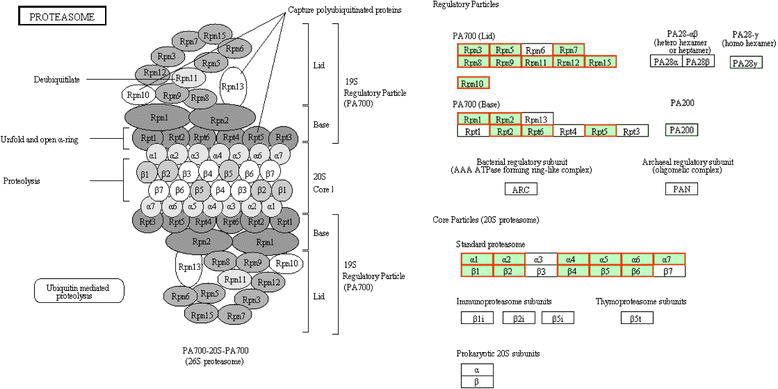
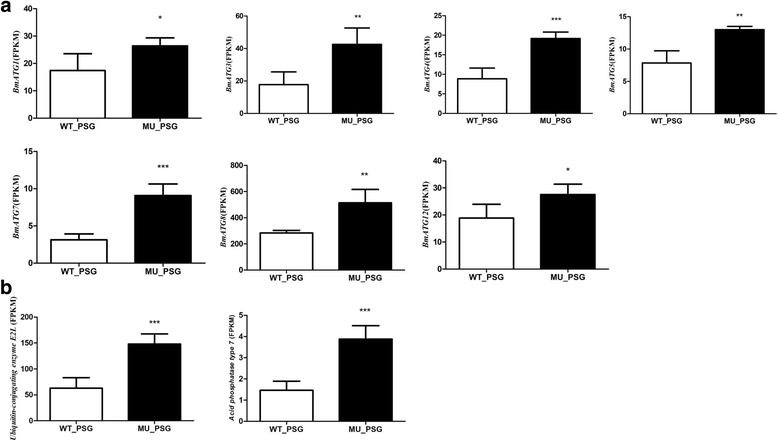
Results: Using CRISPR/Cas9 system, we successfully performed genome editing of Bmfib-H. The loss-of-function mutations caused naked pupa and thin cocoon mutant phenotypes. Compared with the wild type, the posterior silk gland of mutant showed obviously degraded into fragments in advance of programmed cell death of silk gland cells. Comparative transcriptomic analyses of silk gland at the fourth day of the fifth instar larval stage(L5D4)identified 1456 differential expressed genes (DEGs) between posterior silk gland (PSG) and mid silk gland (MSG) and 1388 DEGs between the mutant and the wild type. Hierarchical clustering of all the DEGs indicated a remarkable down-regulated and an up-regulated gene clade in the mutant silk glands, respectively. Down-regulated genes were overrepresented in the pathways involved in cancer, DNA replication and cell proliferation. Intriguingly, up-regulated DEGs are significantly enriched in the proteasome. By further comparison on the transcriptome of MSG and PSG between the wild type and the mutant, we consistently observed that up-regulated DEGs in the mutant PSG were enriched in protein degrading activity and proteasome. Meantime, we observed a series of up-regulated genes involved in autophagy. Since these protein degradation processes would be normally occur after the spinning time, the results suggesting that these progresses were activated remarkably ahead of schedule in the mutant.
Conclusions: Accumulation of abnormal fib-H protein might arouse the activation of proteasomes as well as autophagy process, to promote the rapid degradation of such abnormal proteins and the silk gland cells. Our study therefore proposes a subsequent process of protein and partial cellular degradation caused by mutation of silk protein, which might be helpful for understanding its impact of the silk gland biological process, and further exploration the re-allocation of nitrogen in the silkworm.
Keywords: Autophagy; Bmfib-H; Bombyx mori; CRISPR/Cas9; Proteasome; Transcriptome.
Conflict of interest statement
Ethics approval and consent to participate
The material we used in this study is a significant economic and model lepidopteran insect, the silkworm (
Consent for publication
All the authors have read the manuscript and the additional files and agreed to publish.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
-
- Wang H, Wang Y, Wu C, Tao H, Chen X, Yin W, Sima Y, Wang Y, Xu S. Changes in 30K protein synthesis during delayed degeneration of the silk gland by a caspase-dependent pathway in a Bombyx (silkworm) mutant. J comp physiol B, Biochem, syst, environ physiol. 2016;186(6):689–700. doi: 10.1007/s00360-016-0990-4. - DOI - PubMed
-
- Inoue S, Tanaka K, Arisaka F, Kimura S, Ohtomo K, Mizuno S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6 : 6 : 1 molar ratio. J Biol Chem. 2000;275(51):40517–40528. doi: 10.1074/jbc.M006897200. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

