Performance of in silico prediction tools for the classification of rare BRCA1/2 missense variants in clinical diagnostics
- PMID: 29580235
- PMCID: PMC5870501
- DOI: 10.1186/s12920-018-0353-y
Performance of in silico prediction tools for the classification of rare BRCA1/2 missense variants in clinical diagnostics
Abstract
Background: The use of next-generation sequencing approaches in clinical diagnostics has led to a tremendous increase in data and a vast number of variants of uncertain significance that require interpretation. Therefore, prediction of the effects of missense mutations using in silico tools has become a frequently used approach. Aim of this study was to assess the reliability of in silico prediction as a basis for clinical decision making in the context of hereditary breast and/or ovarian cancer.
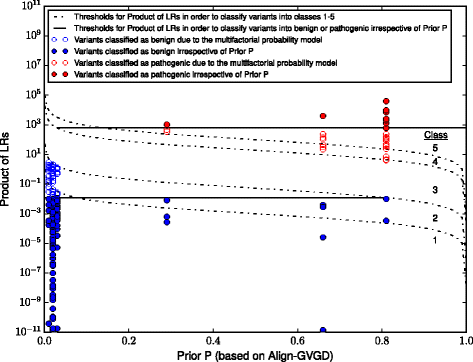
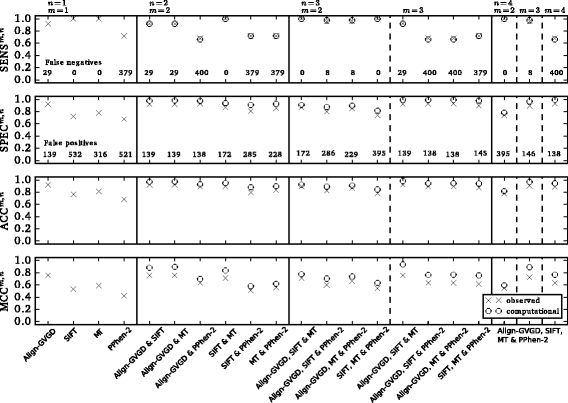
Methods: We tested the performance of four prediction tools (Align-GVGD, SIFT, PolyPhen-2, MutationTaster2) using a set of 236 BRCA1/2 missense variants that had previously been classified by expert committees. However, a major pitfall in the creation of a reliable evaluation set for our purpose is the generally accepted classification of BRCA1/2 missense variants using the multifactorial likelihood model, which is partially based on Align-GVGD results. To overcome this drawback we identified 161 variants whose classification is independent of any previous in silico prediction. In addition to the performance as stand-alone tools we examined the sensitivity, specificity, accuracy and Matthews correlation coefficient (MCC) of combined approaches.
Results: PolyPhen-2 achieved the lowest sensitivity (0.67), specificity (0.67), accuracy (0.67) and MCC (0.39). Align-GVGD achieved the highest values of specificity (0.92), accuracy (0.92) and MCC (0.73), but was outperformed regarding its sensitivity (0.90) by SIFT (1.00) and MutationTaster2 (1.00). All tools suffered from poor specificities, resulting in an unacceptable proportion of false positive results in a clinical setting. This shortcoming could not be bypassed by combination of these tools. In the best case scenario, 138 families would be affected by the misclassification of neutral variants within the cohort of patients of the German Consortium for Hereditary Breast and Ovarian Cancer.
Conclusion: We show that due to low specificities state-of-the-art in silico prediction tools are not suitable to predict pathogenicity of variants of uncertain significance in BRCA1/2. Thus, clinical consequences should never be based solely on in silico forecasts. However, our data suggests that SIFT and MutationTaster2 could be suitable to predict benignity, as both tools did not result in false negative predictions in our analysis.
Keywords: BRCA; Classification; Missense variant; Prediction tools; Variant of uncertain significance.
Conflict of interest statement
Ethics approval and consent to participate
The GC-HBOC registry has been approved by the responsible Ethics Committee of the University of Cologne (07-185, October 18th, 2007). Written informed consent to be enrolled in the GC-HBOC registry, was obtained from all individuals whose data was used for the present study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Byrski T, Dent R, Blecharz P, Foszczynska-Kloda M, Gronwald J, Huzarski T, Cybulski C, Marczyk E, Chrzan R, Eisen A, Lubinski J, Narod SA. Results of a phase II open-label, non-randomized trial of cisplatin chemotherapy in patients with BRCA1-positive metastatic breast cancer. Breast Cancer Res. 2012;14:110. doi: 10.1186/bcr3231. - DOI - PMC - PubMed
-
- Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK, Wardley A, Mitchell G, Earl H, Wickens M, Carmichael J. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. doi: 10.1016/S0140-6736(10)60892-6. - DOI - PubMed
-
- Eccles DM, Mitchell G, Monteiro ANA, Schmutzler R, Couch FJ, Spurdle AB, Gómez-García EB, Driessen R, Lindor NM, Blok MJ, Moller P, de la Hoya M, Pal T, Domchek S, Nathanson K, Van Asperen C, Diez O, Rheim K, Stoppa-Lyonnet D, Parsons M, Goldgar D. BRCA1 and BRCA2 genetic testing–pitfalls and recommendations for managing variants of uncertain clinical significance. Ann Oncol. 2015;26(10):2057–065. doi: 10.1093/annonc/mdv278. - DOI - PMC - PubMed
-
- Moghadasi S, Hofland N, Wouts JN, Hogervorst FBL, Wijnen JT, Vreeswijk MPG, van Asperen CJ. Variants of uncertain significance in BRCA1 and BRCA2 assessment of in silico analysis and a proposal for communication in genetic counselling. J Med Genet. 2013;50(2):74–9. doi: 10.1136/jmedgenet-2012-100961. - DOI - PubMed
-
- Santacroce R, Leccese A, Trunzo R, Lassandro G, Giordano P, Ettorre C, Antoncecchi S, Cantori I, Dragani A, Belvini D, Salviato R, Margaglione M. Identification of ten novel mutations in factor VIII gene: A study of a cohort of 52 haemophilia A patients. Thromb Res. 2015;135(5):1031–4. doi: 10.1016/j.thromres.2015.01.019. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

