Inhibition of anthrax lethal factor by ssDNA aptamers
- PMID: 29580944
- PMCID: PMC5916030
- DOI: 10.1016/j.abb.2018.03.028
Inhibition of anthrax lethal factor by ssDNA aptamers
Abstract
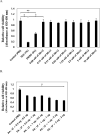
Anthrax is caused by Bacillus anthracis, a bacterium that is able to secrete the toxins protective antigen, edema factor and lethal factor. Due to the high level of secretion from the bacteria and its severe virulence, lethal factor (LF) has been sought as a biomarker for detecting bacterial infection and as an effective target to neutralize toxicity. In this study, we found three aptamers, and binding affinity was determined by fluorescently labeled aptamers. One of the aptamers exhibited high affinity, with a Kd value of 11.0 ± 2.7 nM, along with low cross reactivity relative to bovine serum albumin and protective antigen. The therapeutic functionality of the aptamer was examined by assessing the inhibition of LF protease activity against a mitogen-activated protein kinase kinase. The aptamer appears to be an effective inhibitor of LF with an IC50 value of 15 ± 1.5 μM and approximately 85% cell viability, suggesting that this aptamer provides a potential clue for not only development of a sensitive diagnostic device of B. anthracis infection but also the design of novel inhibitors of LF.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

