HnRNP L represses cryptic exons
- PMID: 29581412
- PMCID: PMC5959245
- DOI: 10.1261/rna.065508.117
HnRNP L represses cryptic exons
Abstract
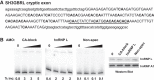
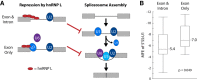
The fidelity of RNA splicing is regulated by a network of splicing enhancers and repressors, although the rules that govern this process are not yet fully understood. One mechanism that contributes to splicing fidelity is the repression of nonconserved cryptic exons by splicing factors that recognize dinucleotide repeats. We previously identified that TDP-43 and PTBP1/PTBP2 are capable of repressing cryptic exons utilizing UG and CU repeats, respectively. Here we demonstrate that hnRNP L (HNRNPL) also represses cryptic exons by utilizing exonic CA repeats, particularly near the 5'SS. We hypothesize that hnRNP L regulates CA repeat repression for both cryptic exon repression and developmental processes such as T cell differentiation.
Keywords: HNRNPL; HnRNP L; alternative splicing; cryptic exons; dinucleotide repeats.
© 2018 McClory et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Ayala YM, Pantano S, D'Ambrogio A, Buratti E, Brindisi A, Marchetti C, Romano M, Baralle FE. 2005. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol 348: 575–588. - PubMed
-
- Blatter M, Dunin-Horkawicz S, Grishina I, Maris C, Thore S, Maier T, Bindereif A, Bujnicki JM, Allain FHT. 2015. The signature of the five-stranded vRRM fold defined by functional, structural and computational analysis of the hnRNP L protein. J Mol Biol 427: 3001–3022. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
