JAM3 maintains leukemia-initiating cell self-renewal through LRP5/AKT/β-catenin/CCND1 signaling
- PMID: 29584620
- PMCID: PMC5919829
- DOI: 10.1172/JCI93198
JAM3 maintains leukemia-initiating cell self-renewal through LRP5/AKT/β-catenin/CCND1 signaling
Abstract
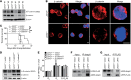
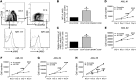
Leukemia-initiating cells (LICs) are responsible for the initiation, development, and relapse of leukemia. The identification of novel therapeutic LIC targets is critical to curing leukemia. In this report, we reveal that junctional adhesion molecule 3 (JAM3) is highly enriched in both mouse and human LICs. Leukemogenesis is almost completely abrogated upon Jam3 deletion during serial transplantations in an MLL-AF9-induced murine acute myeloid leukemia model. In contrast, Jam3 deletion does not affect the functions of mouse hematopoietic stem cells. Moreover, knockdown of JAM3 leads to a dramatic decrease in the proliferation of both human leukemia cell lines and primary LICs. JAM3 directly associates with LRP5 to activate the downstream PDK1/AKT pathway, followed by the downregulation of GSK3β and activation of β-catenin/CCND1 signaling, to maintain the self-renewal ability and cell cycle entry of LICs. Thus, JAM3 may serve as a functional LIC marker and play an important role in the maintenance of LIC stemness through unexpected LRP5/PDK1/AKT/GSK3β/β-catenin/CCND1 signaling pathways but not via its canonical role in cell junctions and migration. JAM3 may be an ideal therapeutic target for the eradication of LICs without influencing normal hematopoiesis.
Keywords: Cancer; Hematology; Leukemias; Stem cells.
Conflict of interest statement
Figures





References
-
- Hutchinson L. Immunotherapy: CAR-modified T cells targeting CD19-curing the incurable. Nat Rev Clin Oncol. 2014;11(12):683. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

