PIKfyve regulates melanosome biogenesis
- PMID: 29584722
- PMCID: PMC5889185
- DOI: 10.1371/journal.pgen.1007290
PIKfyve regulates melanosome biogenesis
Abstract
PIKfyve, VAC14, and FIG4 form a complex that catalyzes the production of PI(3,5)P2, a signaling lipid implicated in process ranging from lysosome maturation to neurodegeneration. While previous studies have identified VAC14 and FIG4 mutations that lead to both neurodegeneration and coat color defects, how PIKfyve regulates melanogenesis is unknown. In this study, we sought to better understand the role of PIKfyve in melanosome biogenesis. Melanocyte-specific PIKfyve knockout mice exhibit greying of the mouse coat and the accumulation of single membrane vesicle structures in melanocytes resembling multivesicular endosomes. PIKfyve inhibition blocks melanosome maturation, the processing of the melanosome protein PMEL, and the trafficking of the melanosome protein TYRP1. Taken together, these studies identify a novel role for PIKfyve in controlling the delivery of proteins from the endosomal compartment to the melanosome, a role that is distinct from the role of PIKfyve in the reformation of lysosomes from endolysosomes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
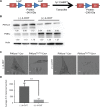
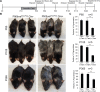

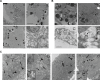


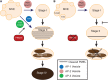
Similar articles
-
The PIKfyve complex regulates the early melanosome homeostasis required for physiological amyloid formation.J Cell Sci. 2019 Feb 28;132(5):jcs229500. doi: 10.1242/jcs.229500. J Cell Sci. 2019. PMID: 30709920 Free PMC article.
-
PIKfyve activity regulates reformation of terminal storage lysosomes from endolysosomes.Traffic. 2017 Nov;18(11):747-757. doi: 10.1111/tra.12525. Traffic. 2017. PMID: 28857423
-
Intermittent inhibition of FYVE finger-containing phosphoinositide kinase induces melanosome degradation in B16F10 melanoma cells.Mol Biol Rep. 2023 Jul;50(7):5917-5930. doi: 10.1007/s11033-023-08536-9. Epub 2023 May 29. Mol Biol Rep. 2023. PMID: 37248430
-
Roles of PIKfyve in multiple cellular pathways.Curr Opin Cell Biol. 2022 Jun;76:102086. doi: 10.1016/j.ceb.2022.102086. Epub 2022 May 16. Curr Opin Cell Biol. 2022. PMID: 35584589 Free PMC article. Review.
-
Mechanisms of protein delivery to melanosomes in pigment cells.Physiology (Bethesda). 2012 Apr;27(2):85-99. doi: 10.1152/physiol.00043.2011. Physiology (Bethesda). 2012. PMID: 22505665 Free PMC article. Review.
Cited by
-
Chemical and biochemical control of skin pigmentation with special emphasis on mixed melanogenesis.Pigment Cell Melanoma Res. 2021 Jul;34(4):730-747. doi: 10.1111/pcmr.12970. Epub 2021 Mar 22. Pigment Cell Melanoma Res. 2021. PMID: 33751833 Free PMC article. Review.
-
The Genetic Background of Abnormalities in Metabolic Pathways of Phosphoinositides and Their Linkage with the Myotubular Myopathies, Neurodegenerative Disorders, and Carcinogenesis.Biomolecules. 2023 Oct 19;13(10):1550. doi: 10.3390/biom13101550. Biomolecules. 2023. PMID: 37892232 Free PMC article. Review.
-
Inhibition of PIKfyve Leads to Lysosomal Disorders via Dysregulation of mTOR Signaling.Cells. 2024 May 30;13(11):953. doi: 10.3390/cells13110953. Cells. 2024. PMID: 38891085 Free PMC article.
-
Recent advances in understanding the molecular basis of melanogenesis in melanocytes.F1000Res. 2020 Jun 15;9:F1000 Faculty Rev-608. doi: 10.12688/f1000research.24625.1. eCollection 2020. F1000Res. 2020. PMID: 32595944 Free PMC article. Review.
-
PIKfyve Deficiency in Myeloid Cells Impairs Lysosomal Homeostasis in Macrophages and Promotes Systemic Inflammation in Mice.Mol Cell Biol. 2019 Oct 11;39(21):e00158-19. doi: 10.1128/MCB.00158-19. Print 2019 Nov 1. Mol Cell Biol. 2019. PMID: 31427458 Free PMC article.
References
-
- Costin GE, Hearing VJ. Human skin pigmentation: melanocytes modulate skin color in response to stress. FASEB J. 2007;21(4):976–94. Epub 2007/01/24. doi: 10.1096/fj.06-6649rev [pii] . - DOI - PubMed
-
- Sitaram A, Marks MS. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology (Bethesda). 2012;27(2):85–99. Epub 2012/04/17. doi: 10.1152/physiol.00043.2011 ; PubMed Central PMCID: PMCPMC3345138. - DOI - PMC - PubMed
-
- Raposo G, Marks MS. Melanosomes—dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol. 2007;8(10):786–97. Epub 2007/09/20. doi: 10.1038/nrm2258 ; PubMed Central PMCID: PMCPMC2786984. - DOI - PMC - PubMed
-
- Wei AH, Li W. Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 2013;26(2):176–92. Epub 2012/11/23. doi: 10.1111/pcmr.12051 . - DOI - PubMed
-
- Wasmeier C, Hume AN, Bolasco G, Seabra MC. Melanosomes at a glance. J Cell Sci. 2008;121(Pt 24):3995–9. Epub 2008/12/06. doi: 10.1242/jcs.040667 [pii]. . - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

