Transcriptome analysis of genetically matched human induced pluripotent stem cells disomic or trisomic for chromosome 21
- PMID: 29584757
- PMCID: PMC5870938
- DOI: 10.1371/journal.pone.0194581
Transcriptome analysis of genetically matched human induced pluripotent stem cells disomic or trisomic for chromosome 21
Abstract
Trisomy of chromosome 21, the genetic cause of Down syndrome, has the potential to alter expression of genes on chromosome 21, as well as other locations throughout the genome. These transcriptome changes are likely to underlie the Down syndrome clinical phenotypes. We have employed RNA-seq to undertake an in-depth analysis of transcriptome changes resulting from trisomy of chromosome 21, using induced pluripotent stem cells (iPSCs) derived from a single individual with Down syndrome. These cells were originally derived by Li et al, who genetically targeted chromosome 21 in trisomic iPSCs, allowing selection of disomic sibling iPSC clones. Analyses were conducted on trisomic/disomic cell pairs maintained as iPSCs or differentiated into cortical neuronal cultures. In addition to characterization of gene expression levels, we have also investigated patterns of RNA adenosine-to-inosine editing, alternative splicing, and repetitive element expression, aspects of the transcriptome that have not been significantly characterized in the context of Down syndrome. We identified significant changes in transcript accumulation associated with chromosome 21 trisomy, as well as changes in alternative splicing and repetitive element transcripts. Unexpectedly, the trisomic iPSCs we characterized expressed higher levels of neuronal transcripts than control disomic iPSCs, and readily differentiated into cortical neurons, in contrast to another reported study. Comparison of our transcriptome data with similar studies of trisomic iPSCs suggests that trisomy of chromosome 21 may not intrinsically limit neuronal differentiation, but instead may interfere with the maintenance of pluripotency.
Conflict of interest statement
Figures
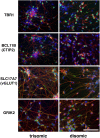


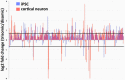



References
-
- Antonarakis SE, Lyle R, Chrast R, Scott HS. Differential gene expression studies to explore the molecular pathophysiology of Down syndrome. Brain Res Brain Res Rev. 2001;36(2–3):265–74. . - PubMed
-
- Mittaz L, Scott HS, Rossier C, Seeburg PH, Higuchi M, Antonarakis SE. Cloning of a human RNA editing deaminase (ADARB1) of glutamate receptors that maps to chromosome 21q22.3. Genomics. 1997;41(2):210–7. doi: 10.1006/geno.1997.4655 . - DOI - PubMed
-
- Lalioti MD, Gos A, Green MR, Rossier C, Morris MA, Antonarakis SE. The gene for human U2 snRNP auxiliary factor small 35-kDa subunit (U2AF1) maps to the progressive myoclonus epilepsy (EPM1) critical region on chromosome 21q22.3. Genomics. 1996;33(2):298–300. doi: 10.1006/geno.1996.0196 . - DOI - PubMed
-
- Shi J, Zhang T, Zhou C, Chohan MO, Gu X, Wegiel J, et al. Increased dosage of Dyrk1A alters alternative splicing factor (ASF)-regulated alternative splicing of tau in Down syndrome. J Biol Chem. 2008;283(42):28660–9. doi: 10.1074/jbc.M802645200 . - DOI - PMC - PubMed
-
- Gardiner K, Herault Y, Lott IT, Antonarakis SE, Reeves RH, Dierssen M. Down syndrome: from understanding the neurobiology to therapy. J Neurosci. 2010;30(45):14943–5. doi: 10.1523/JNEUROSCI.3728-10.2010 . - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

