Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity
- PMID: 29587142
- PMCID: PMC5875457
- DOI: 10.1016/j.devcel.2018.02.024
Developmental History Provides a Roadmap for the Emergence of Tumor Plasticity
Abstract
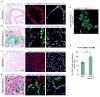
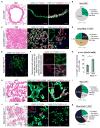
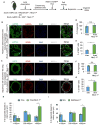
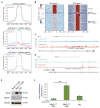
We show that the loss or gain of transcription factor programs that govern embryonic cell-fate specification is associated with a form of tumor plasticity characterized by the acquisition of alternative cell fates normally characteristic of adjacent organs. In human non-small cell lung cancers, downregulation of the lung lineage-specifying TF NKX2-1 is associated with tumors bearing features of various gut tissues. Loss of Nkx2-1 from murine alveolar, but not airway, epithelium results in conversion of lung cells to gastric-like cells. Superimposing oncogenic Kras activation enables further plasticity in both alveolar and airway epithelium, producing tumors that adopt midgut and hindgut fates. Conversely, coupling Nkx2-1 loss with foregut lineage-specifying SOX2 overexpression drives the formation of squamous cancers with features of esophageal differentiation. These findings demonstrate that elements of pathologic tumor plasticity mirror the normal developmental history of organs in that cancer cells acquire cell fates associated with developmentally related neighboring organs.
Keywords: Waddington landscape; developmental history; non-small cell lung cancers; transdifferentiation; tumor heterogeneity; tumor plasticity.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Antonini D, Sirico A, Aberdam E, Ambrosio R, Campanile C, Fagoonee S, Altruda F, Aberdam D, Brissette JL, Missero C. A composite enhancer regulates p63 gene expression in epidermal morphogenesis and in keratinocyte differentiation by multiple mechanisms. Nucleic Acids Res. 2015;43:862–874. - PMC - PubMed
-
- Blanpain C. Tracing the cellular origin of cancer. Nat Cell Biol. 2013;15:126–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

