Spatial-Temporal Lineage Restrictions of Embryonic p63+ Progenitors Establish Distinct Stem Cell Pools in Adult Airways
- PMID: 29587145
- PMCID: PMC5875454
- DOI: 10.1016/j.devcel.2018.03.001
Spatial-Temporal Lineage Restrictions of Embryonic p63+ Progenitors Establish Distinct Stem Cell Pools in Adult Airways
Abstract
Basal cells (BCs) are p63-expressing multipotent progenitors of skin, tracheoesophageal and urinary tracts. p63 is abundant in developing airways; however, it remains largely unclear how embryonic p63+ cells contribute to the developing and postnatal respiratory tract epithelium, and ultimately how they relate to adult BCs. Using lineage-tracing and functional approaches in vivo, we show that p63+ cells arising from the lung primordium are initially multipotent progenitors of airway and alveolar lineages but later become restricted proximally to generate the tracheal adult stem cell pool. In intrapulmonary airways, these cells are maintained immature to adulthood in bronchi, establishing a rare p63+Krt5- progenitor cell population that responds to H1N1 virus-induced severe injury. Intriguingly, this pool includes a CC10 lineage-labeled p63+Krt5- cell subpopulation required for a full H1N1-response. These data elucidate key aspects in the establishment of regionally distinct adult stem cell pools in the respiratory system, potentially with relevance to other organs.
Keywords: H1N1; airway progenitors; basal cells; influenza A; lineage tracing; p63; stem cell.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

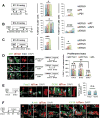
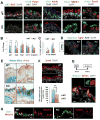
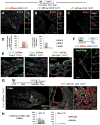

Comment in
-
Basal Cells in Lung Development and Repair.Dev Cell. 2018 Mar 26;44(6):653-654. doi: 10.1016/j.devcel.2018.03.004. Dev Cell. 2018. PMID: 29587138
References
-
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM. Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. Am J Physiol Cell Physiol. 2004;287:C171–C181. - PubMed
-
- Guhar A, Deshpande A, Jain A, Sebastiani, Cardoso WV. Uroplakin 3a+ cells are a distinctive population of epithelial progenitors that contribute to airway maintenance and post-injury repair. Cell Reports. 2017;19:246–254. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

