Assessment of microbiome changes after rumen transfaunation: implications on improving feed efficiency in beef cattle
- PMID: 29587855
- PMCID: PMC5869788
- DOI: 10.1186/s40168-018-0447-y
Assessment of microbiome changes after rumen transfaunation: implications on improving feed efficiency in beef cattle
Abstract
Background: Understanding the host impact on its symbiotic microbiota is important in redirecting the rumen microbiota and thus improving animal performance. The current study aimed to understand how rumen microbiota were altered and re-established after being emptied and receiving content from donor, thus to understand the impact of such process on rumen microbial fermentation and to explore the microbial phylotypes with higher manipulation potentials.
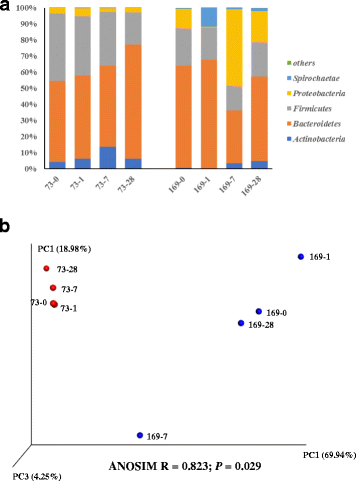
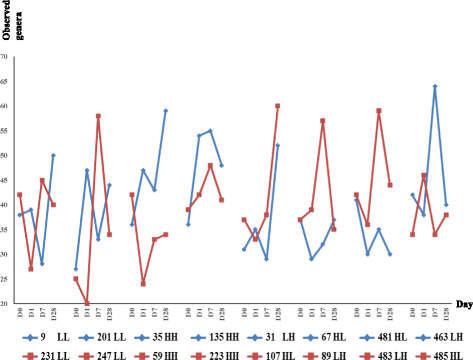
Results: Individual animal had strong effect on the re-establishment of the bacterial community according to the observed profiles detected by both fingerprinting and pyrosequencing. Most of the bacterial profile recovery patterns and extents at genus level varied among steers; and each identified bacterial genus responded to transfaunation differently within each host. Coriobacteriaceae, Coprococcus, and Lactobacillus were found to be the most responsive and tunable genera by exchanging rumen content. Besides, the association of 18 bacterial phylotypes with host fermentation parameters suggest that these phylotypes should also be considered as the regulating targets in improving host feed efficiency. In addition, the archaeal community had different re-establishment patterns for each host as determined by fingerprint profiling: it was altered after receiving non-native microbiome in some animals, while it resumed its original status after the adaptation period in the other ones.
Conclusions: The highly individualized microbial re-establishment process suggested the importance of considering host genetics, microbial functional genomics, and host fermentation/performance assessment when developing effective and selective microbial manipulation methods for improving animal feed efficiency.
Keywords: Adaptation; Beef cattle; Rumen microbiota; Transfaunation.
Conflict of interest statement
Ethics approval
All of the animals used for the current study were obtained from Roy Berg Kinsella Research Ranch, University of Alberta, and were cared for following the guidelines of the Canadian Council of Animal Care (ISBN: 978-0-919087-50-7) (2009, Ottawa, ON, Canada). The animals were transferred to Metabolic Units at Edmonton Research Station, University of Alberta, and the animal study was proved by Animal Care Use Committee Livestock, University of Alberta (Protocol No. AUP00000116).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

