Sequential regulatory activity prediction across chromosomes with convolutional neural networks
- PMID: 29588361
- PMCID: PMC5932613
- DOI: 10.1101/gr.227819.117
Sequential regulatory activity prediction across chromosomes with convolutional neural networks
Abstract
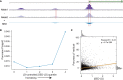
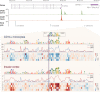
Models for predicting phenotypic outcomes from genotypes have important applications to understanding genomic function and improving human health. Here, we develop a machine-learning system to predict cell-type-specific epigenetic and transcriptional profiles in large mammalian genomes from DNA sequence alone. By use of convolutional neural networks, this system identifies promoters and distal regulatory elements and synthesizes their content to make effective gene expression predictions. We show that model predictions for the influence of genomic variants on gene expression align well to causal variants underlying eQTLs in human populations and can be useful for generating mechanistic hypotheses to enable fine mapping of disease loci.
© 2018 Kelley et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al. 2016. TensorFlow: a system for large-scale machine learning. In USENIX symposium on operating systems design and implementation, Savannah, GA.
-
- Albert FW, Kruglyak L. 2015. The role of regulatory variation in complex traits and disease. Nat Rev Genet 16: 197–212. - PubMed
-
- Alipanahi B, Delong A, Weirauch MT, Frey BJ. 2015. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat Biotechnol 33: 831–838. - PubMed
-
- Ayoubi TA, Van De Ven WJ. 1996. Regulation of gene expression by alternative promoters. FASEB J 10: 453–460. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
