Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles
- PMID: 29588418
- PMCID: PMC5899464
- DOI: 10.1073/pnas.1720542115
Successful reprogramming of cellular protein production through mRNA delivered by functionalized lipid nanoparticles
Abstract
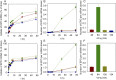
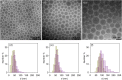
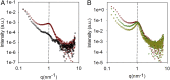
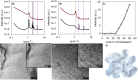
The development of safe and efficacious gene vectors has limited greatly the potential for therapeutic treatments based on messenger RNA (mRNA). Lipid nanoparticles (LNPs) formed by an ionizable cationic lipid (here DLin-MC3-DMA), helper lipids (distearoylphosphatidylcholine, DSPC, and cholesterol), and a poly(ethylene glycol) (PEG) lipid have been identified as very promising delivery vectors of short interfering RNA (siRNA) in different clinical phases; however, delivery of high-molecular weight RNA has been proven much more demanding. Herein we elucidate the structure of hEPO modified mRNA-containing LNPs of different sizes and show how structural differences affect transfection of human adipocytes and hepatocytes, two clinically relevant cell types. Employing small-angle scattering, we demonstrate that LNPs have a disordered inverse hexagonal internal structure with a characteristic distance around 6 nm in presence of mRNA, whereas LNPs containing no mRNA do not display this structure. Furthermore, using contrast variation small-angle neutron scattering, we show that one of the lipid components, DSPC, is localized mainly at the surface of mRNA-containing LNPs. By varying LNP size and surface composition we demonstrate that both size and structure have significant influence on intracellular protein production. As an example, in both human adipocytes and hepatocytes, protein expression levels for 130 nm LNPs can differ as much as 50-fold depending on their surface characteristics, likely due to a difference in the ability of LNP fusion with the early endosome membrane. We consider these discoveries to be fundamental and opening up new possibilities for rational design of synthetic nanoscopic vehicles for mRNA delivery.
Keywords: adipocytes; gene therapy; hEPO mRNA; hepatocytes; small-angle scattering.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
Conflict of interest statement: M.Y.A., T.K., S.B., S.W., X.W., A.J.K, A.D., and L.L. are employed by AstraZeneca R&D Gothenburg.
Figures

References
-
- Kormann MSD, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29:154–157. - PubMed
-
- Yin H, et al. Non-viral vectors for gene-based therapy. Nat Rev Genet. 2014;15:541–555. - PubMed
-
- Dowdy SF. Overcoming cellular barriers for RNA therapeutics. Nat Biotechnol. 2017;35:222–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

