An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease
- PMID: 29589097
- PMCID: PMC5954066
- DOI: 10.1007/s00401-018-1841-z
An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer's disease
Abstract
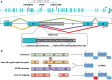
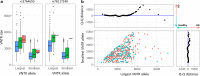
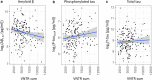
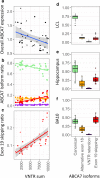
Mutations leading to premature termination codons in ATP-Binding Cassette Subfamily A Member 7 (ABCA7) are high penetrant risk factors of Alzheimer's disease (AD). The influence of other genetic variants in ABCA7 and downstream functional mechanisms, however, is poorly understood. To address this knowledge gap, we investigated tandem repetitive regions in ABCA7 in a Belgian cohort of 1529 AD patients and control individuals and identified an intronic variable number tandem repeat (VNTR). We observed strong association between VNTR length and a genome-wide associated signal for AD in the ABCA7 locus. Expanded VNTR alleles were highly enriched in AD patients [odds ratio = 4.5 (1.3-24.2)], and VNTR length inversely correlated with amyloid β1-42 in cerebrospinal fluid and ABCA7 expression. In addition, we identified three novel ABCA7 alternative splicing events. One isoform in particular-which is formed through exon 19 skipping-lacks the first nucleotide binding domain of ABCA7 and is abundant in brain tissue. We observed a tight correlation between exon 19 skipping and VNTR length. Our findings underline the importance of studying repetitive DNA in complex disorders and expand the contribution of genetic and transcript variation in ABCA7 to AD.
Keywords: ATP-Binding Cassette, Sub-Family A, Member 7 (ABCA7); Alternative splicing; Alzheimer’s disease; Cerebrospinal fluid (CSF) biomarkers; Variable number tandem repeat (VNTR).
Conflict of interest statement
Informed consent
All participants and/or their legal guardian gave written informed consent for participation in clinical and genetic studies. Autopsied patients or their legal guardian gave written informed consent for inclusion in neuropathological studies. Clinical study protocol and the informed consent forms for patient ascertainment were approved by the ethic committee of the respective hospitals at the cohort sampling sites. The genetic study protocols and informed consent forms were approved by the Ethics Committees of the University of Antwerp and the University Hospital of Antwerp, Belgium.
Conflict of interest
The authors declare no conflict of interest.
Figures
References
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

