Chemically induced proximity in biology and medicine
- PMID: 29590011
- PMCID: PMC6417506
- DOI: 10.1126/science.aao5902
Chemically induced proximity in biology and medicine
Abstract
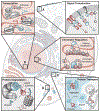
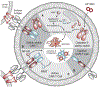
Proximity, or the physical closeness of molecules, is a pervasive regulatory mechanism in biology. For example, most posttranslational modifications such as phosphorylation, methylation, and acetylation promote proximity of molecules to play deterministic roles in cellular processes. To understand the role of proximity in biologic mechanisms, chemical inducers of proximity (CIPs) were developed to synthetically model biologically regulated recruitment. Chemically induced proximity allows for precise temporal control of transcription, signaling cascades, chromatin regulation, protein folding, localization, and degradation, as well as a host of other biologic processes. A systematic analysis of CIPs in basic research, coupled with recent technological advances utilizing CRISPR, distinguishes roles of causality from coincidence and allows for mathematical modeling in synthetic biology. Recently, induced proximity has provided new avenues of gene therapy and emerging advances in cancer treatment.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures



References
-
- Hänggi P, Talkner P, Borkovec M, Reaction-rate theory: Fifty years after Kramers. Rev. Mod. Phys 62, 251–341 (1990). doi: 10.1103/RevModPhys.62.251 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

