NF90/ILF3 is a transcription factor that promotes proliferation over differentiation by hierarchical regulation in K562 erythroleukemia cells
- PMID: 29590119
- PMCID: PMC5873942
- DOI: 10.1371/journal.pone.0193126
NF90/ILF3 is a transcription factor that promotes proliferation over differentiation by hierarchical regulation in K562 erythroleukemia cells
Abstract
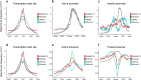
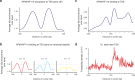
NF90 and splice variant NF110 are DNA- and RNA-binding proteins encoded by the Interleukin enhancer-binding factor 3 (ILF3) gene that have been established to regulate RNA splicing, stabilization and export. The roles of NF90 and NF110 in regulating transcription as chromatin-interacting proteins have not been comprehensively characterized. Here, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) identified 9,081 genomic sites specifically occupied by NF90/NF110 in K562 cells. One third of NF90/NF110 peaks occurred at promoters of annotated genes. NF90/NF110 occupancy colocalized with chromatin marks associated with active promoters and strong enhancers. Comparison with 150 ENCODE ChIP-seq experiments revealed that NF90/NF110 clustered with transcription factors exhibiting preference for promoters over enhancers (POLR2A, MYC, YY1). Differential gene expression analysis following shRNA knockdown of NF90/NF110 in K562 cells revealed that NF90/NF110 activates transcription factors that drive growth and proliferation (EGR1, MYC), while attenuating differentiation along the erythroid lineage (KLF1). NF90/NF110 associates with chromatin to hierarchically regulate transcription factors that promote proliferation and suppress differentiation.
Conflict of interest statement
Figures




Similar articles
-
Inducible expression of immediate early genes is regulated through dynamic chromatin association by NF45/ILF2 and NF90/NF110/ILF3.PLoS One. 2019 Apr 25;14(4):e0216042. doi: 10.1371/journal.pone.0216042. eCollection 2019. PLoS One. 2019. PMID: 31022259 Free PMC article.
-
NF45 and NF90/NF110 coordinately regulate ESC pluripotency and differentiation.RNA. 2017 Aug;23(8):1270-1284. doi: 10.1261/rna.061499.117. Epub 2017 May 9. RNA. 2017. PMID: 28487382 Free PMC article.
-
Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection.Mol Cell. 2017 Jul 20;67(2):214-227.e7. doi: 10.1016/j.molcel.2017.05.023. Epub 2017 Jun 15. Mol Cell. 2017. PMID: 28625552
-
Ilf3 and NF90 functions in RNA biology.Wiley Interdiscip Rev RNA. 2015 Mar-Apr;6(2):243-56. doi: 10.1002/wrna.1270. Epub 2014 Oct 18. Wiley Interdiscip Rev RNA. 2015. PMID: 25327818 Review.
-
NF90 isoforms, a new family of cellular proteins involved in viral replication?Biochimie. 2015 Jan;108:20-4. doi: 10.1016/j.biochi.2014.10.022. Epub 2014 Nov 4. Biochimie. 2015. PMID: 25447144 Review.
Cited by
-
Insights Into the Involvement of Circular RNAs in Autoimmune Diseases.Front Immunol. 2021 Feb 25;12:622316. doi: 10.3389/fimmu.2021.622316. eCollection 2021. Front Immunol. 2021. PMID: 33717126 Free PMC article. Review.
-
The role of circular RNAs in autoimmune diseases: Potential diagnostic biomarkers and therapeutic targets.FASEB J. 2025 Jan 31;39(2):e70263. doi: 10.1096/fj.202401764R. FASEB J. 2025. PMID: 39873909 Free PMC article. Review.
-
DNA and RNA Binding Proteins: From Motifs to Roles in Cancer.Int J Mol Sci. 2022 Aug 18;23(16):9329. doi: 10.3390/ijms23169329. Int J Mol Sci. 2022. PMID: 36012592 Free PMC article. Review.
-
Remodeling of the endothelial cell transcriptional program via paracrine and DNA-binding activities of MPO.iScience. 2024 Jan 12;27(2):108898. doi: 10.1016/j.isci.2024.108898. eCollection 2024 Feb 16. iScience. 2024. PMID: 38322992 Free PMC article.
-
NF90 stabilizes cyclin E1 mRNA through phosphorylation of NF90-Ser382 by CDK2.Cell Death Discov. 2020 Jan 22;6:3. doi: 10.1038/s41420-020-0236-9. eCollection 2020. Cell Death Discov. 2020. PMID: 32123579 Free PMC article.
References
-
- Kornberg RD. Eukaryotic transcriptional control. Trends Cell Biol. 1999;9(12):M46–9. - PubMed
-
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424(6945):147–51. doi: 10.1038/nature01763 - DOI - PubMed
-
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128(4):635–8. doi: 10.1016/j.cell.2007.02.006 - DOI - PubMed
-
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33 Suppl:245–54. - PubMed
-
- Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

