Comprehensive Molecular Profiling Identifies FOXM1 as a Key Transcription Factor for Meningioma Proliferation
- PMID: 29590631
- PMCID: PMC8204688
- DOI: 10.1016/j.celrep.2018.03.013
Comprehensive Molecular Profiling Identifies FOXM1 as a Key Transcription Factor for Meningioma Proliferation
Abstract
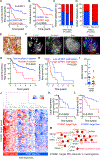
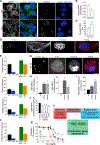
Meningioma is the most common primary intracranial tumor, but the molecular drivers of aggressive meningioma are incompletely understood. Using 280 human meningioma samples and RNA sequencing, immunohistochemistry, whole-exome sequencing, DNA methylation arrays, and targeted gene expression profiling, we comprehensively define the molecular profile of aggressive meningioma. Transcriptomic analyses identify FOXM1 as a key transcription factor for meningioma proliferation and a marker of poor clinical outcomes. Consistently, we discover genomic and epigenomic factors associated with FOXM1 activation in aggressive meningiomas. Finally, we define a FOXM1/Wnt signaling axis in meningioma that is associated with a mitotic gene expression program, poor clinical outcomes, and proliferation of primary meningioma cells. In summary, we find that multiple molecular mechanisms converge on a FOXM1/Wnt signaling axis in aggressive meningioma.
Keywords: DNA methylation; FOXM1; NF2; RNA sequencing; Wnt; epigenome; genome; meningioma; transcriptome; whole-exome sequencing.
Published by Elsevier Inc.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures
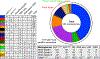
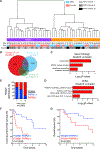
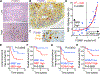
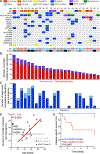

References
-
- Bi WL, Abedalthagafi M, Horowitz P, Agarwalla PK, Mei Y, Aizer AA, Brewster R, Dunn GP, Al-Mefty O, Alexander BM, et al. (2016). Genomic landscape of intracranial meningiomas. J. Neurosurg 125, 525–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

