Disease Ontology: improving and unifying disease annotations across species
- PMID: 29590633
- PMCID: PMC5897730
- DOI: 10.1242/dmm.032839
Disease Ontology: improving and unifying disease annotations across species
Abstract
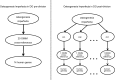
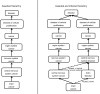
Model organisms are vital to uncovering the mechanisms of human disease and developing new therapeutic tools. Researchers collecting and integrating relevant model organism and/or human data often apply disparate terminologies (vocabularies and ontologies), making comparisons and inferences difficult. A unified disease ontology is required that connects data annotated using diverse disease terminologies, and in which the terminology relationships are continuously maintained. The Mouse Genome Database (MGD, http://www.informatics.jax.org), Rat Genome Database (RGD, http://rgd.mcw.edu) and Disease Ontology (DO, http://www.disease-ontology.org) projects are collaborating to augment DO, aligning and incorporating disease terms used by MGD and RGD, and improving DO as a tool for unifying disease annotations across species. Coordinated assessment of MGD's and RGD's disease term annotations identified new terms that enhance DO's representation of human diseases. Expansion of DO term content and cross-references to clinical vocabularies (e.g. OMIM, ORDO, MeSH) has enriched the DO's domain coverage and utility for annotating many types of data generated from experimental and clinical investigations. The extension of anatomy-based DO classification structure of disease improves accessibility of terms and facilitates application of DO for computational research. A consistent representation of disease associations across data types from cellular to whole organism, generated from clinical and model organism studies, will promote the integration, mining and comparative analysis of these data. The coordinated enrichment of the DO and adoption of DO by MGD and RGD demonstrates DO's usability across human data, MGD, RGD and the rest of the model organism database community.
Keywords: Disease models; Mouse; Ontologies; Rat.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

