Neutrophil-specific knockout demonstrates a role for mitochondria in regulating neutrophil motility in zebrafish
- PMID: 29590639
- PMCID: PMC5897731
- DOI: 10.1242/dmm.033027
Neutrophil-specific knockout demonstrates a role for mitochondria in regulating neutrophil motility in zebrafish
Abstract
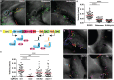
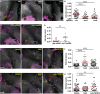
Neutrophils are fast-moving cells essential for host immune functions. Although they primarily rely on glycolysis for ATP, isolated primary human neutrophils depend on mitochondrial membrane potential for chemotaxis. However, it is not known whether mitochondria regulate neutrophil motility in vivo, and the underlying molecular mechanisms remain obscure. Here, we visualized mitochondria in an interconnected network that localizes to the front and rear of migrating neutrophils using a novel transgenic zebrafish line. To disrupt mitochondrial function genetically, we established a gateway system harboring the CRISPR/Cas9 elements for tissue-specific knockout. In a transgenic line, neutrophil-specific disruption of mitochondrial DNA polymerase, polg, significantly reduced the velocity of neutrophil interstitial migration. In addition, inhibiting the mitochondrial electron transport chain or the enzymes that reduce mitochondrial reactive oxygen species also inhibited neutrophil motility. The reduced cell motility that resulted from neutrophil-specific knockout of sod1 was rescued with sod1 mRNA overexpression, or by treating with scavengers of reactive oxygen species. Together, our work has provided the first in vivo evidence that mitochondria regulate neutrophil motility, as well as tools for the functional characterization of mitochondria-related genes in neutrophils and insights into immune deficiency seen in patients with primary mitochondrial disorders.This article has an associated First Person interview with the first author of the paper.
Keywords: Cell migration; Mitochondria; Neutrophil; Tissue-specific knockout; Zebrafish.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

