Discovery of physiological and cancer-related regulators of 3' UTR processing with KAPAC
- PMID: 29592812
- PMCID: PMC5875010
- DOI: 10.1186/s13059-018-1415-3
Discovery of physiological and cancer-related regulators of 3' UTR processing with KAPAC
Abstract
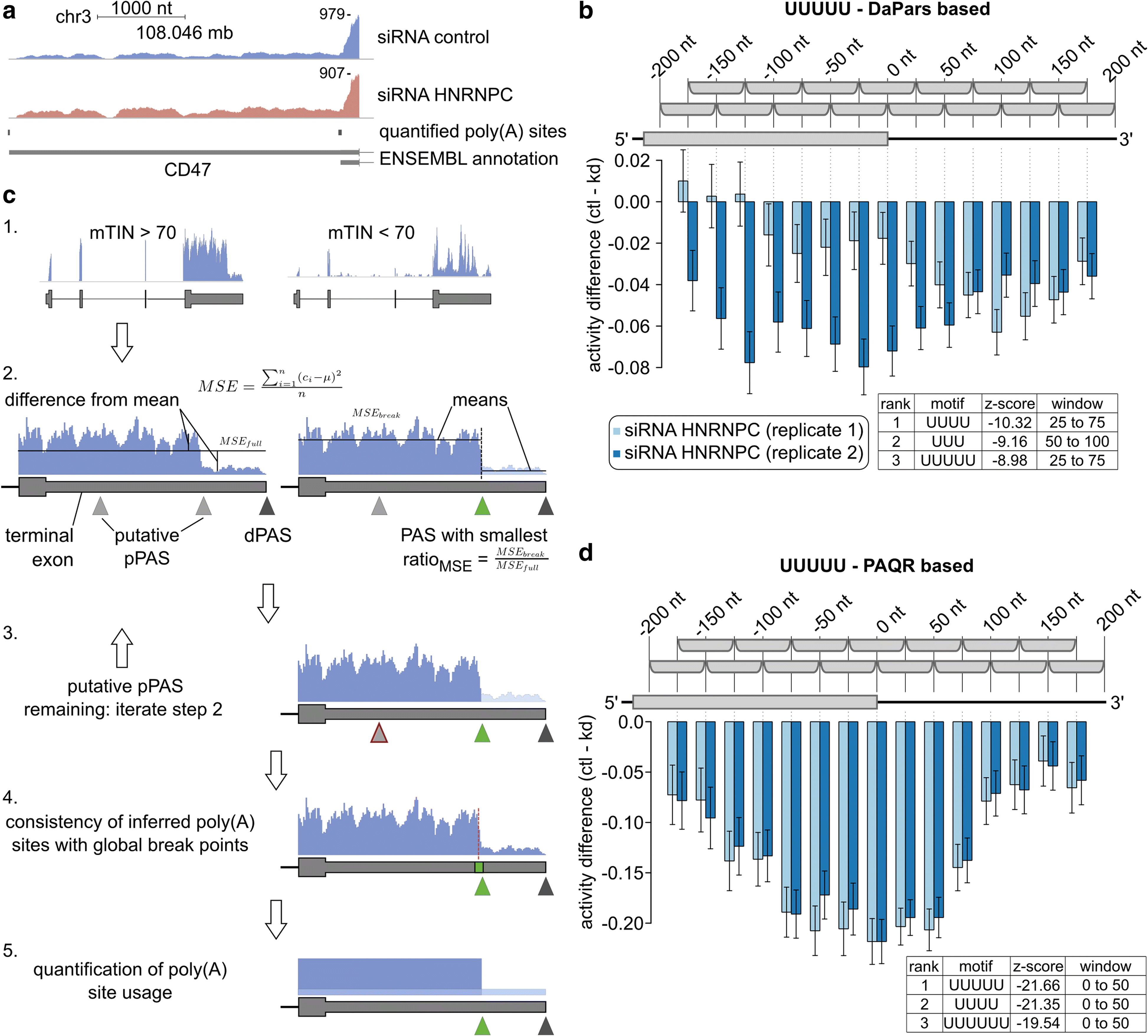
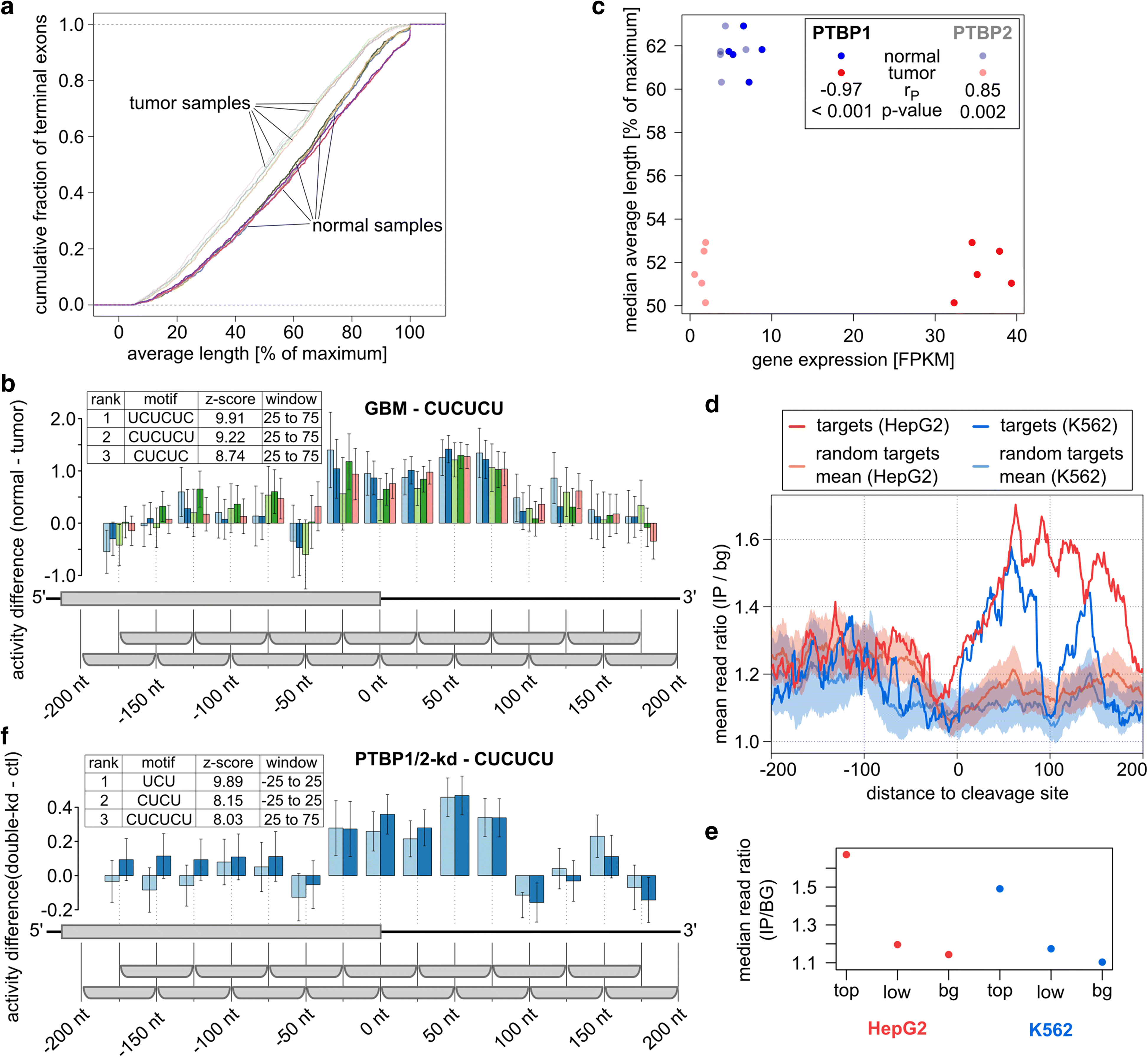
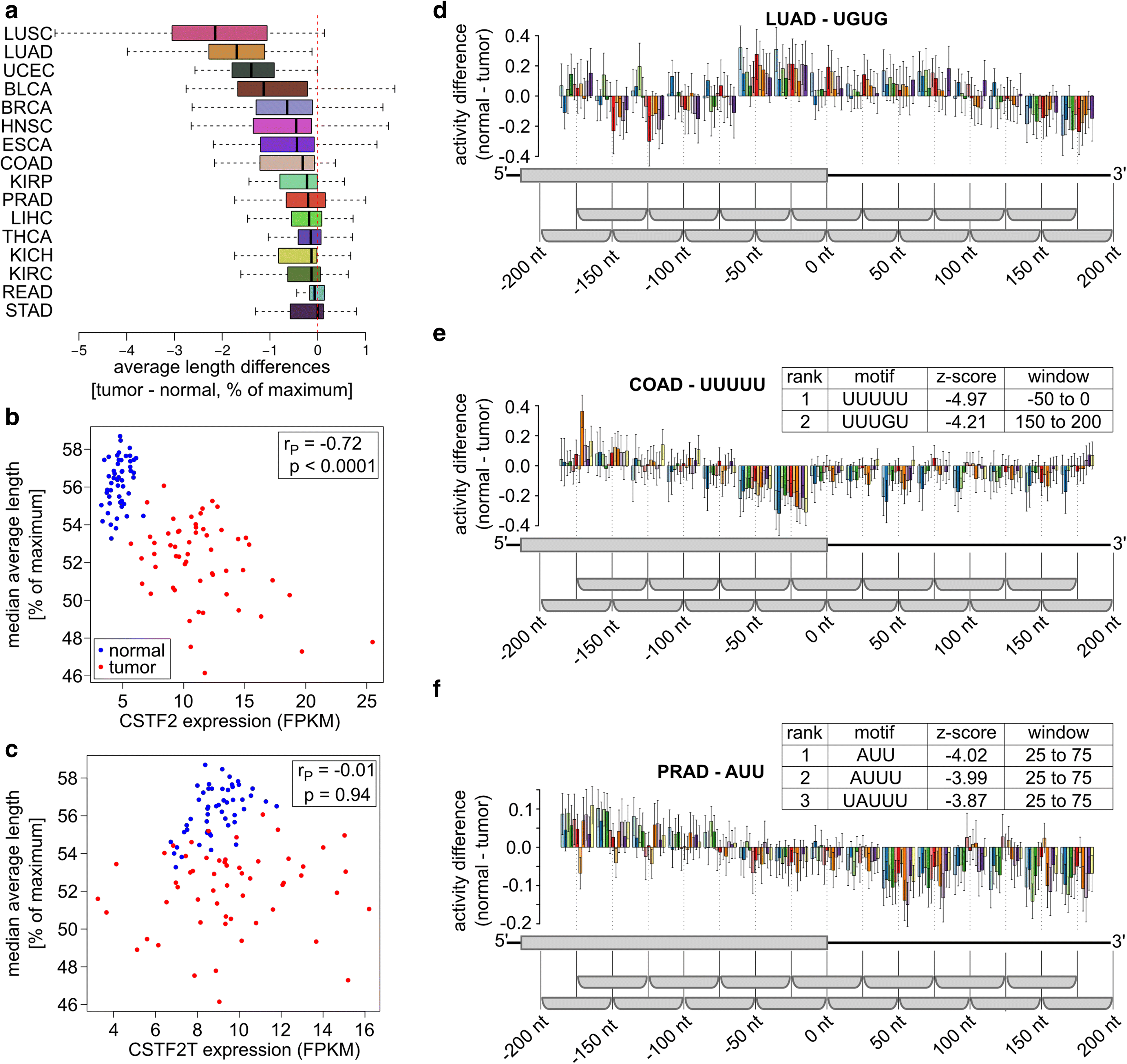
3' Untranslated regions (3' UTRs) length is regulated in relation to cellular state. To uncover key regulators of poly(A) site use in specific conditions, we have developed PAQR, a method for quantifying poly(A) site use from RNA sequencing data and KAPAC, an approach that infers activities of oligomeric sequence motifs on poly(A) site choice. Application of PAQR and KAPAC to RNA sequencing data from normal and tumor tissue samples uncovers motifs that can explain changes in cleavage and polyadenylation in specific cancers. In particular, our analysis points to polypyrimidine tract binding protein 1 as a regulator of poly(A) site choice in glioblastoma.
Keywords: APA; CFIm; Cleavage and polyadenylation; Colon adenocarcinoma; Glioblastoma; HNRNPC; KAPAC; PAQR; PTBP1; Prostate adenocarcinoma.
Conflict of interest statement
Ethics approval and consent to participate
Authorization to use RNA-seq data from patient samples, which is obtained by TCGA Research Network, has been granted.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

