Decreased A-to-I RNA editing as a source of keratinocytes' dsRNA in psoriasis
- PMID: 29592874
- PMCID: PMC5959251
- DOI: 10.1261/rna.064659.117
Decreased A-to-I RNA editing as a source of keratinocytes' dsRNA in psoriasis
Abstract
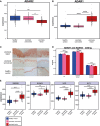
Recognition of dsRNA molecules activates the MDA5-MAVS pathway and plays a critical role in stimulating type-I interferon responses in psoriasis. However, the source of the dsRNA accumulation in psoriatic keratinocytes remains largely unknown. A-to-I RNA editing is a common co- or post-transcriptional modification that diversifies adenosine in dsRNA, and leads to unwinding of dsRNA structures. Thus, impaired RNA editing activity can result in an increased load of endogenous dsRNAs. Here we provide a transcriptome-wide analysis of RNA editing across dozens of psoriasis patients, and we demonstrate a global editing reduction in psoriatic lesions. In addition to the global alteration, we also detect editing changes in functional recoding sites located in the IGFBP7, COPA, and FLNA genes. Accretion of dsRNA activates autoimmune responses, and therefore the results presented here, linking for the first time an autoimmune disease to reduction in global editing level, are relevant to a wide range of autoimmune diseases.
Keywords: A-to-I; RNA editing; interferon; psoriasis.
© 2018 Shallev et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures





References
-
- Andrews S. 2010. FastQC: A quality control tool for high throughput sequence data. Vol. 2011. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
