An APOO Pseudogene on Chromosome 5q Is Associated With Low-Density Lipoprotein Cholesterol Levels
- PMID: 29593015
- PMCID: PMC6162188
- DOI: 10.1161/CIRCULATIONAHA.118.034016
An APOO Pseudogene on Chromosome 5q Is Associated With Low-Density Lipoprotein Cholesterol Levels
Abstract
Background: Elevated levels of low-density lipoprotein cholesterol (LDL-C) are a major risk factor for cardiovascular disease via its contribution to the development and progression of atherosclerotic lesions. Although the genetic basis of LDL-C has been studied extensively, currently known genetic variants account for only ≈20% of the variation in LDL-C levels.
Methods: Through an array-based association analysis in 1102 Amish subjects, we identified a variant strongly associated with LDL-C levels. Using a combination of genetic analyses, zebrafish models, and in vitro experiments, we sought to identify the causal gene driving this association.
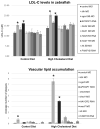
Results: We identified a founder haplotype associated with a 15 mg/dL increase in LDL-C on chromosome 5. After recombination mapping, the associated region contained 8 candidate genes. Using a zebrafish model to evaluate the relevance of these genes to cholesterol metabolism, we found that expression of the transcribed pseudogene, APOOP1, increased LDL-C and vascular plaque formation.
Conclusions: Based on these data, we propose that APOOP1 regulates levels of LDL-C in humans, thus identifying a novel mechanism of lipid homeostasis.
Keywords: cholesterol, LDL; chromosome mapping; founder effect; genetics; pseudogenes.
Figures



Similar articles
-
The prevalence and correlates of subclinical atherosclerosis among adults with low-density lipoprotein cholesterol <70 mg/dL: The Multi-Ethnic Study of Atherosclerosis (MESA) and Brazilian Longitudinal Study of Adult Health (ELSA-Brasil).Atherosclerosis. 2018 Jul;274:61-66. doi: 10.1016/j.atherosclerosis.2018.04.021. Epub 2018 Apr 28. Atherosclerosis. 2018. PMID: 29751286
-
Association of apolipoprotein E polymorphism with plasma lipid disorders, independent of obesity-related traits in Vietnamese children.Lipids Health Dis. 2016 Oct 10;15(1):176. doi: 10.1186/s12944-016-0349-6. Lipids Health Dis. 2016. PMID: 27724906 Free PMC article.
-
APOC3 Loss-of-Function Mutations, Remnant Cholesterol, Low-Density Lipoprotein Cholesterol, and Cardiovascular Risk: Mediation- and Meta-Analyses of 137 895 Individuals.Arterioscler Thromb Vasc Biol. 2018 Mar;38(3):660-668. doi: 10.1161/ATVBAHA.117.310473. Epub 2018 Jan 18. Arterioscler Thromb Vasc Biol. 2018. PMID: 29348120
-
Association of m.5178C>A variant with serum lipid levels: a systematic review and meta-analysis.Biosci Rep. 2021 Dec 22;41(12):BSR20212246. doi: 10.1042/BSR20212246. Biosci Rep. 2021. PMID: 34859818 Free PMC article.
-
Surprises From Genetic Analyses of Lipid Risk Factors for Atherosclerosis.Circ Res. 2016 Feb 19;118(4):579-85. doi: 10.1161/CIRCRESAHA.115.306398. Circ Res. 2016. PMID: 26892959 Free PMC article. Review.
Cited by
-
Pseudogenes in Cardiovascular Disease.Front Mol Biosci. 2021 Feb 10;7:622540. doi: 10.3389/fmolb.2020.622540. eCollection 2020. Front Mol Biosci. 2021. PMID: 33644114 Free PMC article. Review.
-
Proteomic Analysis and 2-Hydroxyisobutyrylation Profiling in Metabolic Syndrome Induced Restenosis.Mol Cell Proteomics. 2025 Jun;24(6):100978. doi: 10.1016/j.mcpro.2025.100978. Epub 2025 Apr 24. Mol Cell Proteomics. 2025. PMID: 40287094 Free PMC article.
-
Apolipoprotein O modulates cholesterol metabolism via NRF2/CYB5R3 independent of LDL receptor.Cell Death Dis. 2024 Jun 3;15(6):389. doi: 10.1038/s41419-024-06778-4. Cell Death Dis. 2024. PMID: 38830896 Free PMC article.
-
An Amish founder population reveals rare-population genetic determinants of the human lipidome.Commun Biol. 2022 Apr 7;5(1):334. doi: 10.1038/s42003-022-03291-2. Commun Biol. 2022. PMID: 35393526 Free PMC article.
-
Prevalence, control, and treatment of diabetes, hypertension, and high cholesterol in the Amish.BMJ Open Diabetes Res Care. 2020 Aug;8(1):e000912. doi: 10.1136/bmjdrc-2019-000912. BMJ Open Diabetes Res Care. 2020. PMID: 32843497 Free PMC article.
References
-
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans. The San Antonio Family Heart Study. Circulation. 1996;94:2159–2170. - PubMed
-
- Rice T, Vogler GP, Perry TS, Laskarzewski PM, Rao DC. Familial aggregation of lipids and lipoproteins in families ascertained through random and nonrandom probands in the Iowa Lipid Research Clinics family study. Hum Hered. 1991;41:107–121. - PubMed
-
- Pollin TI, Hsueh WC, Steinle NI, Snitker S, Shuldiner AR, Mitchell BD. A genome-wide scan of serum lipid levels in the Old Order Amish. Atherosclerosis. 2004;173:89–96. - PubMed
-
- de Ferranti SD, Rodday AM, Mendelson MM, Wong JB, Leslie LK, Sheldrick RC. Prevalence of familial hypercholesterolemia in the 1999 to 2012 United States National Health and Nutrition Examination Surveys (NHANES) Circulation. 2016;133:1067–1072. - PubMed
-
- Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, Kettunen J, Pirinen M, Karjalainen J, Thorleifsson G, Hagg S, Hottenga JJ, Isaacs A, Ladenvall C, Beekman M, Esko T, Ried JS, Nelson CP, Willenborg C, Gustafsson S, Westra HJ, Blades M, de Craen AJ, de Geus EJ, Deelen J, Grallert H, Hamsten A, Havulinna AS, Hengstenberg C, Houwing-Duistermaat JJ, Hypponen E, Karssen LC, Lehtimaki T, Lyssenko V, Magnusson PK, Mihailov E, Muller-Nurasyid M, Mpindi JP, Pedersen NL, Penninx BW, Perola M, Pers TH, Peters A, Rung J, Smit JH, Steinthorsdottir V, Tobin MD, Tsernikova N, van Leeuwen EM, Viikari JS, Willems SM, Willemsen G, Schunkert H, Erdmann J, Samani NJ, Kaprio J, Lind L, Gieger C, Metspalu A, Slagboom PE, Groop L, van Duijn CM, Eriksson JG, Jula A, Salomaa V, Boomsma DI, Power C, Raitakari OT, Ingelsson E, Jarvelin MR, Thorsteinsdottir U, Franke L, Ikonen E, Kallioniemi O, Pietiainen V, Lindgren CM, Stefansson K, Palotie A, McCarthy MI, Morris AP, Prokopenko I, Ripatti S. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

