eCD4-Ig Variants That More Potently Neutralize HIV-1
- PMID: 29593050
- PMCID: PMC5974481
- DOI: 10.1128/JVI.02011-17
eCD4-Ig Variants That More Potently Neutralize HIV-1
Abstract
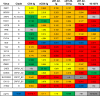
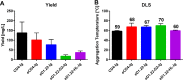
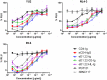
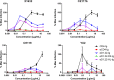
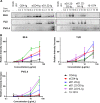
The human immunodeficiency virus type 1 (HIV-1) entry inhibitor eCD4-Ig is a fusion of CD4-Ig and a coreceptor-mimetic peptide. eCD4-Ig is markedly more potent than CD4-Ig, with neutralization efficiencies approaching those of HIV-1 broadly neutralizing antibodies (bNAbs). However, unlike bNAbs, eCD4-Ig neutralized all HIV-1, HIV-2, and simian immunodeficiency virus (SIV) isolates that it has been tested against, suggesting that it may be useful in clinical settings, where antibody escape is a concern. Here, we characterize three new eCD4-Ig variants, each with a different architecture and each utilizing D1.22, a stabilized form of CD4 domain 1. These variants were 10- to 20-fold more potent than our original eCD4-Ig variant, with a construct bearing four D1.22 domains (eD1.22-HL-Ig) exhibiting the greatest potency. However, this variant mediated less efficient antibody-dependent cell-mediated cytotoxicity (ADCC) activity than eCD4-Ig itself or several other eCD4-Ig variants, including the smallest variant (eD1.22-Ig). A variant with the same architecture as the original eCD4-Ig (eD1.22-D2-Ig) showed modestly higher thermal stability and best prevented the promotion of infection of CCR5-positive, CD4-negative cells. All three variants, and eCD4-Ig itself, mediated more efficient shedding of the HIV-1 envelope glycoprotein gp120 than did CD4-Ig. Finally, we show that only three D1.22 mutations contributed to the potency of eD1.22-D2-Ig and that introduction of these changes into eCD4-Ig resulted in a variant 9-fold more potent than eCD4-Ig and 2-fold more potent than eD1.22-D2-Ig. These studies will assist in developing eCD4-Ig variants with properties optimized for prophylaxis, therapy, and cure applications.IMPORTANCE HIV-1 bNAbs have properties different from those of antiretroviral compounds. Specifically, antibodies can enlist immune effector cells to eliminate infected cells, whereas antiretroviral compounds simply interfere with various steps in the viral life cycle. Unfortunately, HIV-1 is adept at evading antibody recognition, limiting the utility of antibodies as a treatment for HIV-1 infection or as part of an effort to eradicate latently infected cells. eCD4-Ig is an antibody-like entry inhibitor that closely mimics HIV-1's obligate receptors. eCD4-Ig appears to be qualitatively different from antibodies, since it neutralizes all HIV-1, HIV-2, and SIV isolates. Here, we characterize three new structurally distinct eCD4-Ig variants and show that each excels in a key property useful to prevent, treat, or cure an HIV-1 infection. For example, one variant neutralized HIV-1 most efficiently, while others best enlisted natural killer cells to eliminate infected cells. These observations will help generate eCD4-Ig variants optimized for different clinical applications.
Keywords: CD4; CD4-Ig; coreceptor; eCD4-Ig; human immunodeficiency virus.
Copyright © 2018 American Society for Microbiology.
Figures
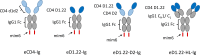


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

