A Strategy To Isolate Modifiers of Caenorhabditis elegans Lethal Mutations: Investigating the Endoderm Specifying Ability of the Intestinal Differentiation GATA Factor ELT-2
- PMID: 29593072
- PMCID: PMC5940137
- DOI: 10.1534/g3.118.200079
A Strategy To Isolate Modifiers of Caenorhabditis elegans Lethal Mutations: Investigating the Endoderm Specifying Ability of the Intestinal Differentiation GATA Factor ELT-2
Abstract
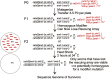
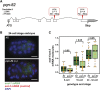
The ELT-2 GATA factor normally functions in differentiation of the C. elegans endoderm, downstream of endoderm specification. We have previously shown that, if ELT-2 is expressed sufficiently early, it is also able to specify the endoderm and to replace all other members of the core GATA-factor transcriptional cascade (END-1, END-3, ELT-7). However, such rescue requires multiple copies (and presumably overexpression) of the end-1p::elt-2 cDNA transgene; a single copy of the transgene does not rescue. We have made this observation the basis of a genetic screen to search for genetic modifiers that allow a single copy of the end-1p::elt-2 cDNA transgene to rescue the lethality of the end-1 end-3 double mutant. We performed this screen on a strain that has a single copy insertion of the transgene in an end-1 end-3 background. These animals are kept alive by virtue of an extrachromosomal array containing multiple copies of the rescuing transgene; the extrachromosomal array also contains a toxin under heat shock control to counterselect for mutagenized survivors that have been able to lose the rescuing array. A screen of ∼14,000 mutagenized haploid genomes produced 17 independent surviving strains. Whole genome sequencing was performed to identify genes that incurred independent mutations in more than one surviving strain. The C. elegans gene tasp-1 was mutated in four independent strains. tasp-1 encodes the C. elegans homolog of Taspase, a threonine-aspartic acid protease that has been found, in both mammals and insects, to cleave several proteins involved in transcription, in particular MLL1/trithorax and TFIIA. A second gene, pqn-82, was mutated in two independent strains and encodes a glutamine-asparagine rich protein. tasp-1 and pqn-82 were verified as loss-of-function modifiers of the end-1p::elt-2 transgene by RNAi and by CRISPR/Cas9-induced mutations. In both cases, gene loss leads to modest increases in the level of ELT-2 protein in the early endoderm although ELT-2 levels do not strictly correlate with rescue. We suggest that tasp-1 and pqn-82 represent a class of genes acting in the early embryo to modulate levels of critical transcription factors or to modulate the responsiveness of critical target genes. The screen's design, rescuing lethality with an extrachromosomal transgene followed by counterselection, has a background survival rate of <10-4 without mutagenesis and should be readily adapted to the general problem of identifying suppressors of C. elegans lethal mutations.
Keywords: C. elegans; ELT-2 GATA factor; Mutant Screen Report; endoderm specification; intestine; pqn-82; tasp-1; taspase.
Copyright © 2018 Wiesenfahrt et al.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
