A conserved transcriptional response to intranasal Ebola virus exposure in nonhuman primates prior to onset of fever
- PMID: 29593102
- PMCID: PMC9986849
- DOI: 10.1126/scitranslmed.aaq1016
A conserved transcriptional response to intranasal Ebola virus exposure in nonhuman primates prior to onset of fever
Abstract
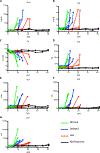
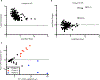
Ebola virus disease (EVD), caused by Ebola virus (EBOV), is a severe illness characterized by case fatality rates of up to 90%. The sporadic nature of outbreaks in resource-limited areas has hindered the ability to characterize the pathogenesis of EVD at all stages of infection but particularly early host responses. Pathogenesis is often studied in nonhuman primate (NHP) models of disease that replicate major aspects of human EVD. Typically, NHP models use a large infectious dose, are carried out through intramuscular or aerosol exposure, and have a fairly uniform disease course. By contrast, we report our analysis of the host response to EBOV after intranasal exposure. Twelve cynomolgus macaques were infected with 100 plaque-forming units of EBOV/Makona through intranasal exposure and presented with varying times to onset of EVD. We used RNA sequencing and a newly developed NanoString CodeSet to monitor the host response via changes in RNA transcripts over time. When individual animal gene expression data were phased based on the onset of sustained fever, the first clinical sign of severe disease, mathematical models indicated that interferon-stimulated genes appeared as early as 4 days before fever onset. This demonstrates that lethal EVD has a uniform and predictable response to infection regardless of time to onset. Furthermore, expression of a subset of genes could predict disease development before other host-based indications of infection such as fever.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures





References
-
- Center for Disease Control (CDC), 2014. Ebola Outbreak in West Africa - Case Counts | Ebola Hemorrhagic Fever | CDC (available at https://www.cdc.gov/vhf/ebola/outbreaks/2014-west-africa/case-counts.html).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

