Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China
- PMID: 29593254
- PMCID: PMC5874244
- DOI: 10.1038/s41426-018-0048-7
Phylogenetic analysis and virulence determinant of the host-adapted Staphylococcus aureus lineage ST188 in China
Abstract
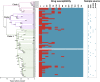
Staphylococcus aureus (S. aureus) is an important pathogen of humans and livestock species, but an understanding of the clonal distribution of S. aureus causing different host-species infections in the same geographical environment and within the same period is lacking. By characterizing infections caused by S. aureus in bovine, pediatric, and adult patients in Shanghai, China, between 2012 and 2014, we identified methicillin-sensitive S. aureus (MSSA) ST188 as the major lineage causing infections in multiple host species. Whole-genome sequencing and phenotypic analyses demonstrated that ST188 might evolve from livestock, and there was no significant genomic or virulence difference between ST188 isolated from livestock and humans. The virulence of ST188 is related to its adhesion and nasal colonization ability. This result is in accord with the strong epithelial cell adhesion and biofilm formation properties of ST188. Furthermore, the adhesion- and biofilm-formation-related genes are present in multiple copies and exhibit significantly increased expression in ST188. In conclusion, S. aureus ST188 is the major lineage causing human and livestock infections in Shanghai, China. Due to its high expression of the factors associated with bacterial adhesion and biofilm formation, ST188 has the ability to colonize and infect different host species.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
