Distinct ECG Phenotypes Identified in Hypertrophic Cardiomyopathy Using Machine Learning Associate With Arrhythmic Risk Markers
- PMID: 29593570
- PMCID: PMC5859357
- DOI: 10.3389/fphys.2018.00213
Distinct ECG Phenotypes Identified in Hypertrophic Cardiomyopathy Using Machine Learning Associate With Arrhythmic Risk Markers
Abstract
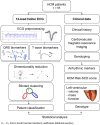
Aims: Ventricular arrhythmia triggers sudden cardiac death (SCD) in hypertrophic cardiomyopathy (HCM), yet electrophysiological biomarkers are not used for risk stratification. Our aim was to identify distinct HCM phenotypes based on ECG computational analysis, and characterize differences in clinical risk factors and anatomical differences using cardiac magnetic resonance (CMR) imaging. Methods: High-fidelity 12-lead Holter ECGs from 85 HCM patients and 38 healthy volunteers were analyzed using mathematical modeling and computational clustering to identify phenotypic subgroups. Clinical features and the extent and distribution of hypertrophy assessed by CMR were evaluated in the subgroups. Results: QRS morphology alone was crucial to identify three HCM phenotypes with very distinct QRS patterns. Group 1 (n = 44) showed normal QRS morphology, Group 2 (n = 19) showed short R and deep S waves in V4, and Group 3 (n = 22) exhibited short R and long S waves in V4-6, and left QRS axis deviation. However, no differences in arrhythmic risk or distribution of hypertrophy were observed between these groups. Including T wave biomarkers in the clustering, four HCM phenotypes were identified: Group 1A (n = 20), with primary repolarization abnormalities showing normal QRS yet inverted T waves, Group 1B (n = 24), with normal QRS morphology and upright T waves, and Group 2 and Group 3 remaining as before, with upright T waves. Group 1A patients, with normal QRS and inverted T wave, showed increased HCM Risk-SCD scores (1A: 4.0%, 1B: 1.8%, 2: 2.1%, 3: 2.5%, p = 0.0001), and a predominance of coexisting septal and apical hypertrophy (p < 0.0001). HCM patients in Groups 2 and 3 exhibited predominantly septal hypertrophy (85 and 90%, respectively). Conclusion: HCM patients were classified in four subgroups with distinct ECG features. Patients with primary T wave inversion not secondary to QRS abnormalities had increased HCM Risk-SCD scores and coexisting septal and apical hypertrophy, suggesting that primary T wave inversion may increase SCD risk in HCM, rather than T wave inversion secondary to depolarization abnormalities. Computational ECG phenotyping provides insight into the underlying processes captured by the ECG and has the potential to be a novel and independent factor for risk stratification.
Keywords: computational clustering; e-cardiology; electrocardiography; hypertrophic cardiomyopathy; phenotyping.
Figures



References
-
- Adabag A. S., Maron B. J., Appelbaum E., Harrigan C. J., Buros J. L., Gibson C. M., et al. (2008). Occurrence and frequency of arrhythmias in hypertrophic cardiomyopathy in relation to delayed enhancement on cardiovascular magnetic resonance. J. Am. Coll. Cardiol. 51, 1369–1374. 10.1016/j.jacc.2007.11.071 - DOI - PubMed
-
- Belkin M., Niyogi P. (2003). Laplacian eigenmaps for dimensionality reduction and data representation. Neural Comput. 15, 1373–1396. 10.1162/089976603321780317 - DOI
-
- Cai D., Zhang C., He X. (2010). Unsupervised feature selection for multi-cluster data, in Proceedings of the 16th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining [Internet] (New York, NY: ACM; ) (cited June 10, 2016), 333–342. Available online at: http://doi.acm.org/10.1145/1835804.1835848 - DOI
-
- Cerqueira M. D., Weissman N. J., Dilsizian V., Jacobs A. K., Kaul S., Laskey W. K., et al. (2002). Standardized myocardial segmentation and nomenclature for tomographic imaging of the heart. A statement for healthcare professionals from the Cardiac imaging committee of the council on clinical Cardiology of the American Heart Association. Circulation 105, 539–542. 10.1161/hc0402.102975 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

