Metabolic Adaptation of Methanogens in Anaerobic Digesters Upon Trace Element Limitation
- PMID: 29593674
- PMCID: PMC5859356
- DOI: 10.3389/fmicb.2018.00405
Metabolic Adaptation of Methanogens in Anaerobic Digesters Upon Trace Element Limitation
Abstract
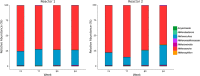
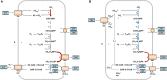
Anaerobic digestion (AD) is a complex multi-stage process relying on the activity of highly diverse microbial communities including hydrolytic, acidogenic and syntrophic acetogenic bacteria as well as methanogenic archaea. The lower diversity of methanogenic archaea compared to the bacterial groups involved in AD and the corresponding lack of functional redundancy cause a stronger susceptibility of methanogenesis to unfavorable process conditions such as trace element (TE) deprivation, thus controlling the stability of the overall process. Here, we investigated the effects of a slowly increasing TE deficit on the methanogenic community function in a semi-continuous biogas process. The aim of the study was to understand how methanogens in digester communities cope with TE limitation and sustain their growth and metabolic activity. Two lab-scale biogas reactors fed with distillers grains and supplemented with TEs were operated in parallel for 76 weeks before one of the reactors was subjected to TE deprivation, leading to a decline of cobalt and molybdenum concentrations from 0.9 to 0.2 mg/L, nickel concentrations from 2.9 to 0.8 mg/L, manganese concentrations from 38 to 18 mg/L, and tungsten concentrations from 1.4 to 0.2 mg/L. Amplicon sequencing of mcrA genes revealed Methanosarcina (72%) and Methanoculleus (23%) as the predominant methanogens in the undisturbed reactors. With increasing TE limitation, the relative abundance of Methanosarcina dropped to 67% and a slight decrease of acetoclastic methanogenic activity was observed in batch tests with 13C-methyl-labeled acetate, suggesting a shift toward syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis. Metaproteome analysis revealed abundance shifts of the enzymes involved in methanogenic pathways. Proteins involved in methylotrophic and acetoclastic methanogenesis decreased in abundance while formylmethanofuran dehydrogenase from Methanosarcinaceae increased, confirming our hypothesis of a shift from acetoclastic to hydrogenotrophic methanogenesis by Methanosarcina. Both Methanosarcina and Methanoculleus increased the abundance of N5-methyltetrahydromethanopterin-coenzyme M methyltransferase and methyl-coenzyme M reductase. However, these efforts to preserve the ion motive force for energy conservation were seemingly more successful in Methanoculleus. We conclude that both methanogenic genera use different strategies to stabilize their energy balance under TE limitation. Methanosarcina switched from TE expensive pathways (methylotrophic and acetoclastic methanogenesis) to hydrogenotrophic methanogenesis. Methanoculleus showed a higher robustness and was favored over the more fastidious Methanosarcina, thus stabilizing reactor performance under TE limitation.
Keywords: Methanoculleus; Methanosarcina; biogas process; mcrA; metaproteome; methanogenic pathways; trace metals.
Figures

References
-
- Ariunbaatar J., Esposito G., Yeh D. H., Lens P. N. L. (2016). Enhanced anaerobic digestion of food waste by supplementing trace elements: role of selenium (VI) and iron (II). Front. Environ. Sci. 4:8 10.3389/fenvs.2016.00008 - DOI
-
- Burke S. A., Krzycki J. A. (1997). Reconstitution of monomethylamine:coenzyme M methyl transfer with a corrinoid protein and two methyltransferases purified from Methanosarcina barkeri. J. Biol. Chem. 272, 16570–16577. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

