Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci
- PMID: 29593713
- PMCID: PMC5859386
- DOI: 10.3389/fimmu.2018.00425
Variable Extent of Lineage-Specificity and Developmental Stage-Specificity of Cohesin and CCCTC-Binding Factor Binding Within the Immunoglobulin and T Cell Receptor Loci
Abstract
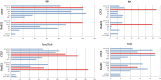
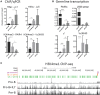
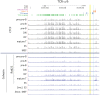
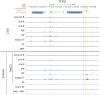
CCCTC-binding factor (CTCF) is largely responsible for the 3D architecture of the genome, in concert with the action of cohesin, through the creation of long-range chromatin loops. Cohesin is hypothesized to be the main driver of these long-range chromatin interactions by the process of loop extrusion. Here, we performed ChIP-seq for CTCF and cohesin in two stages each of T and B cell differentiation and examined the binding pattern in all six antigen receptor (AgR) loci in these lymphocyte progenitors and in mature T and B cells, ES cells, and fibroblasts. The four large AgR loci have many bound CTCF sites, most of which are only occupied in lymphocytes, while only the CTCF sites at the end of each locus near the enhancers or J genes tend to be bound in non-lymphoid cells also. However, despite the generalized lymphocyte restriction of CTCF binding in AgR loci, the Igκ locus is the only locus that also shows significant lineage-specificity (T vs. B cells) and developmental stage-specificity (pre-B vs. pro-B) in CTCF binding. We show that cohesin binding shows greater lineage- and stage-specificity than CTCF at most AgR loci, providing more specificity to the loops. We also show that the culture of pro-B cells in IL7, a common practice to expand the number of cells before ChIP-seq, results in a CTCF-binding pattern resembling pre-B cells, as well as other epigenetic and transcriptional characteristics of pre-B cells. Analysis of the orientation of the CTCF sites show that all sites within the large V portions of the Igh and TCRβ loci have the same orientation. This suggests either a lack of requirement for convergent CTCF sites creating loops, or indicates an absence of any loops between CTCF sites within the V region portion of those loci but only loops to the convergent sites at the D-J-enhancer end of each locus. The V region portions of the Igκ and TCRα/δ loci, by contrast, have CTCF sites in both orientations, providing many options for creating CTCF-mediated convergent loops throughout the loci. CTCF/cohesin loops, along with transcription factors, drives contraction of AgR loci to facilitate the creation of a diverse repertoire of antibodies and T cell receptors.
Keywords: 3D chromatin topology; CCCTC-binding factor; T cell receptor loci; cohesin; immunoglobulin loci; long-range looping.
Figures






Similar articles
-
Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development.J Immunol. 2009 Jan 1;182(1):44-8. doi: 10.4049/jimmunol.182.1.44. J Immunol. 2009. PMID: 19109133 Free PMC article.
-
CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells.Proc Natl Acad Sci U S A. 2011 Jun 7;108(23):9566-71. doi: 10.1073/pnas.1019391108. Epub 2011 May 23. Proc Natl Acad Sci U S A. 2011. PMID: 21606361 Free PMC article.
-
Genome Topology Control of Antigen Receptor Gene Assembly.J Immunol. 2020 May 15;204(10):2617-2626. doi: 10.4049/jimmunol.1901356. J Immunol. 2020. PMID: 32366683 Free PMC article. Review.
-
CTCF orchestrates long-range cohesin-driven V(D)J recombinational scanning.Nature. 2020 Oct;586(7828):305-310. doi: 10.1038/s41586-020-2578-0. Epub 2020 Jul 27. Nature. 2020. PMID: 32717742 Free PMC article.
-
CTCF as a boundary factor for cohesin-mediated loop extrusion: evidence for a multi-step mechanism.Nucleus. 2020 Dec;11(1):132-148. doi: 10.1080/19491034.2020.1782024. Nucleus. 2020. PMID: 32631111 Free PMC article. Review.
Cited by
-
Two Successive Inversional Vβ Rearrangements on a Single Tcrb Allele Can Contribute to the TCRβ Repertoire.J Immunol. 2020 Jan 1;204(1):78-86. doi: 10.4049/jimmunol.1901105. Epub 2019 Nov 18. J Immunol. 2020. PMID: 31740488 Free PMC article.
-
Dynamic 3D Locus Organization and Its Drivers Underpin Immunoglobulin Recombination.Front Immunol. 2021 Feb 18;11:633705. doi: 10.3389/fimmu.2020.633705. eCollection 2020. Front Immunol. 2021. PMID: 33679727 Free PMC article. Review.
-
Endogenous topoisomerase II-mediated DNA breaks drive thymic cancer predisposition linked to ATM deficiency.Nat Commun. 2020 Feb 14;11(1):910. doi: 10.1038/s41467-020-14638-w. Nat Commun. 2020. PMID: 32060399 Free PMC article.
-
Igh and Igk loci use different folding principles for V gene recombination due to distinct chromosomal architectures of pro-B and pre-B cells.Nat Commun. 2023 Apr 21;14(1):2316. doi: 10.1038/s41467-023-37994-9. Nat Commun. 2023. PMID: 37085514 Free PMC article.
-
Enhancers as regulators of antigen receptor loci three-dimensional chromatin structure.Transcription. 2020 Feb;11(1):37-51. doi: 10.1080/21541264.2019.1699383. Epub 2019 Dec 12. Transcription. 2020. PMID: 31829768 Free PMC article. Review.
References
-
- Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol (1996) 16:2802–13.10.1128/MCB.16.6.2802 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

