Orotidine 5'-Monophosphate Decarboxylase: Probing the Limits of the Possible for Enzyme Catalysis
- PMID: 29595949
- PMCID: PMC6016548
- DOI: 10.1021/acs.accounts.8b00059
Orotidine 5'-Monophosphate Decarboxylase: Probing the Limits of the Possible for Enzyme Catalysis
Abstract
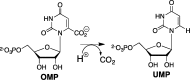
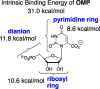
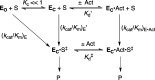
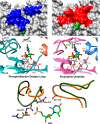
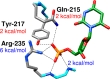
The mystery associated with catalysis by what were once regarded as protein black boxes, diminished with the X-ray crystallographic determination of the three-dimensional structures of enzyme-substrate complexes. The report that several high-resolution X-ray crystal structures of orotidine 5'-monophosphate decarboxylase (OMPDC) failed to provide a consensus mechanism for enzyme-catalyzed decarboxylation of OMP to form uridine 5'-monophosphate, therefore, provoked a flurry of controversy. This controversy was fueled by the enormous 1023-fold rate acceleration for this enzyme, which had " jolted many biochemists' assumptions about the catalytic potential of enzymes." Our studies on the mechanism of action of OMPDC provide strong evidence that catalysis by this enzyme is not fundamentally different from less proficient catalysts, while highlighting important architectural elements that enable a peak level of performance. Many enzymes undergo substrate-induced protein conformational changes that trap their substrates in solvent occluded protein cages, but the conformational change induced by ligand binding to OMPDC is incredibly complex, as required to enable the development of 22 kcal/mol of stabilizing binding interactions with the phosphodianion and ribosyl substrate fragments of OMP. The binding energy from these fragments is utilized to activate OMPDC for catalysis of decarboxylation at the orotate fragment of OMP, through the creation of a tight, catalytically active, protein cage from the floppy, open, unliganded form of OMPDC. Such utilization of binding energy for ligand-driven conformational changes provides a general mechanism to obtain specificity in transition state binding. The rate enhancement that results from the binding of carbon acid substrates to enzymes is partly due to a reduction in the carbon acid p Ka that is associated with ligand binding. The binding of UMP to OMPDC results in an unusually large >12 unit decrease in the p Ka = 29 for abstraction of the C-6 substrate hydrogen, due to stabilization of an enzyme-bound vinyl carbanion, which is also an intermediate of OMPDC-catalyzed decarboxylation. The protein-ligand interactions operate to stabilize the vinyl carbanion at the enzyme active site compared to aqueous solution, rather than to stabilize the transition state for the concerted electrophilic displacement of CO2 by H+ that avoids formation of this reaction intermediate. There is evidence that OMPDC induces strain into the bound substrate. The interaction between the amide side chain of Gln-215 from the phosphodianion gripper loop and the hydroxymethylene side chain of Ser-154 from the pyrimidine umbrella of ScOMPDC position the amide side chain to interact with the phosphodianion of OMP. There are no direct stabilizing interactions between dianion gripper protein side chains Gln-215, Tyr-217, and Arg-235 and the pyrimidine ring at the decarboxylation transition state. Rather these side chains function solely to hold OMPDC in the catalytically active closed conformation. The hydrophobic side chains that line the active site of OMPDC in the region of the departing CO2 product may function to stabilize the decarboxylation transition state by providing hydrophobic solvation of this product.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
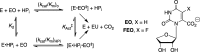
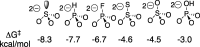






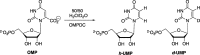




Similar articles
-
Role of the Carboxylate in Enzyme-Catalyzed Decarboxylation of Orotidine 5'-Monophosphate: Transition State Stabilization Dominates Over Ground State Destabilization.J Am Chem Soc. 2019 Aug 28;141(34):13468-13478. doi: 10.1021/jacs.9b04823. Epub 2019 Aug 14. J Am Chem Soc. 2019. PMID: 31365243 Free PMC article.
-
Linear Free Energy Relationships for Enzymatic Reactions: Fresh Insight from a Venerable Probe.Acc Chem Res. 2021 May 18;54(10):2532-2542. doi: 10.1021/acs.accounts.1c00147. Epub 2021 May 3. Acc Chem Res. 2021. PMID: 33939414 Free PMC article. Review.
-
Role of a guanidinium cation-phosphodianion pair in stabilizing the vinyl carbanion intermediate of orotidine 5'-phosphate decarboxylase-catalyzed reactions.Biochemistry. 2013 Oct 22;52(42):7500-11. doi: 10.1021/bi401117y. Epub 2013 Oct 8. Biochemistry. 2013. PMID: 24053466 Free PMC article.
-
Protein-Ribofuranosyl Interactions Activate Orotidine 5'-Monophosphate Decarboxylase for Catalysis.Biochemistry. 2021 Nov 16;60(45):3362-3373. doi: 10.1021/acs.biochem.1c00589. Epub 2021 Nov 2. Biochemistry. 2021. PMID: 34726391 Free PMC article.
-
Specificity in transition state binding: the Pauling model revisited.Biochemistry. 2013 Mar 26;52(12):2021-35. doi: 10.1021/bi301491r. Epub 2013 Feb 4. Biochemistry. 2013. PMID: 23327224 Free PMC article. Review.
Cited by
-
Modeling the Role of a Flexible Loop and Active Site Side Chains in Hydride Transfer Catalyzed by Glycerol-3-phosphate Dehydrogenase.ACS Catal. 2020 Oct 2;10(19):11253-11267. doi: 10.1021/acscatal.0c02757. Epub 2020 Sep 3. ACS Catal. 2020. PMID: 33042609 Free PMC article.
-
Kinetics and mechanism for enzyme-catalyzed reactions of substrate pieces.Methods Enzymol. 2023;685:95-126. doi: 10.1016/bs.mie.2023.03.002. Epub 2023 Apr 18. Methods Enzymol. 2023. PMID: 37245916 Free PMC article.
-
Role of the Carboxylate in Enzyme-Catalyzed Decarboxylation of Orotidine 5'-Monophosphate: Transition State Stabilization Dominates Over Ground State Destabilization.J Am Chem Soc. 2019 Aug 28;141(34):13468-13478. doi: 10.1021/jacs.9b04823. Epub 2019 Aug 14. J Am Chem Soc. 2019. PMID: 31365243 Free PMC article.
-
Orotidine 5'-Monophosphate Decarboxylase: The Operation of Active Site Chains Within and Across Protein Subunits.Biochemistry. 2020 Jun 2;59(21):2032-2040. doi: 10.1021/acs.biochem.0c00241. Epub 2020 May 19. Biochemistry. 2020. PMID: 32374983 Free PMC article.
-
Linear Free Energy Relationships for Enzymatic Reactions: Fresh Insight from a Venerable Probe.Acc Chem Res. 2021 May 18;54(10):2532-2542. doi: 10.1021/acs.accounts.1c00147. Epub 2021 May 3. Acc Chem Res. 2021. PMID: 33939414 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

