Machine-learned analysis of the association of next-generation sequencing-based human TRPV1 and TRPA1 genotypes with the sensitivity to heat stimuli and topically applied capsaicin
- PMID: 29596157
- PMCID: PMC6012053
- DOI: 10.1097/j.pain.0000000000001222
Machine-learned analysis of the association of next-generation sequencing-based human TRPV1 and TRPA1 genotypes with the sensitivity to heat stimuli and topically applied capsaicin
Abstract
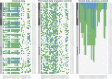
Heat pain and its modulation by capsaicin varies among subjects in experimental and clinical settings. A plausible cause is a genetic component, of which TRPV1 ion channels, by their response to both heat and capsaicin, are primary candidates. However, TRPA1 channels can heterodimerize with TRPV1 channels and carry genetic variants reported to modulate heat pain sensitivity. To address the role of these candidate genes in capsaicin-induced hypersensitization to heat, pain thresholds acquired before and after topical application of capsaicin and TRPA1/TRPV1 exomic sequences derived by next-generation sequencing were assessed in n = 75 healthy volunteers and the genetic information comprised 278 loci. Gaussian mixture modeling indicated 2 phenotype groups with high or low capsaicin-induced hypersensitization to heat. Unsupervised machine learning implemented as swarm-based clustering hinted at differences in the genetic pattern between these phenotype groups. Several methods of supervised machine learning implemented as random forests, adaptive boosting, k-nearest neighbors, naive Bayes, support vector machines, and for comparison, binary logistic regression predicted the phenotype group association consistently better when based on the observed genotypes than when using a random permutation of the exomic sequences. Of note, TRPA1 variants were more important for correct phenotype group association than TRPV1 variants. This indicates a role of the TRPA1 and TRPV1 next-generation sequencing-based genetic pattern in the modulation of the individual response to heat-related pain phenotypes. When considering earlier evidence that topical capsaicin can induce neuropathy-like quantitative sensory testing patterns in healthy subjects, implications for future analgesic treatments with transient receptor potential inhibitors arise.
Conflict of interest statement
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Figures
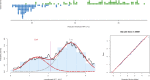
 and shown as probability density function (PDF) estimated by means of the Pareto density estimation (PDE; black line) overlaid on a histogram could be fitted using a Gaussian mixture model (GMM) given as
and shown as probability density function (PDF) estimated by means of the Pareto density estimation (PDE; black line) overlaid on a histogram could be fitted using a Gaussian mixture model (GMM) given as , with M = 2 modes. The fit is shown as a red line and the M = 2 mixes are indicated as differently colored dashed lines (G #1–#2). The Bayesian boundary between the Gaussians is indicated as a perpendicular magenta line. At the right side, a quantile–quantile (QQ) plot is shown comparing the observed distribution of cold pain data (ordinate) with the distribution expected from the GMM (abscissa). The blue dots symbolize the quantiles of observed data vs predicted data and the red line indicates identity, ie, the agreement between the data distribution expected from the model with the observed data distribution. The close vicinity of the dots to this line indicates satisfactory fits of the data by the respective GMM. The figure has been created using the R software package (version 3.4.1 for Linux;
, with M = 2 modes. The fit is shown as a red line and the M = 2 mixes are indicated as differently colored dashed lines (G #1–#2). The Bayesian boundary between the Gaussians is indicated as a perpendicular magenta line. At the right side, a quantile–quantile (QQ) plot is shown comparing the observed distribution of cold pain data (ordinate) with the distribution expected from the GMM (abscissa). The blue dots symbolize the quantiles of observed data vs predicted data and the red line indicates identity, ie, the agreement between the data distribution expected from the model with the observed data distribution. The close vicinity of the dots to this line indicates satisfactory fits of the data by the respective GMM. The figure has been created using the R software package (version 3.4.1 for Linux; 


Similar articles
-
Machine-Learned Association of Next-Generation Sequencing-Derived Variants in Thermosensitive Ion Channels Genes with Human Thermal Pain Sensitivity Phenotypes.Int J Mol Sci. 2020 Jun 19;21(12):4367. doi: 10.3390/ijms21124367. Int J Mol Sci. 2020. PMID: 32575443 Free PMC article.
-
TRPA1 Sensitization Produces Hyperalgesia to Heat but not to Cold Stimuli in Human Volunteers.Clin J Pain. 2019 Apr;35(4):321-327. doi: 10.1097/AJP.0000000000000677. Clin J Pain. 2019. PMID: 30664549 Clinical Trial.
-
Psychophysical and vasomotor evidence for interdependency of TRPA1 and TRPV1-evoked nociceptive responses in human skin: an experimental study.Pain. 2018 Oct;159(10):1989-2001. doi: 10.1097/j.pain.0000000000001298. Pain. 2018. PMID: 29847470
-
Is thermal nociception only sensed by the capsaicin receptor, TRPV1?Anat Sci Int. 2009 Sep;84(3):122-8. doi: 10.1007/s12565-009-0048-8. Epub 2009 Jun 27. Anat Sci Int. 2009. PMID: 19562439 Review.
-
Targeting TRP channels for pain relief: A review of current evidence from bench to bedside.Curr Opin Pharmacol. 2024 Apr;75:102447. doi: 10.1016/j.coph.2024.102447. Epub 2024 Mar 11. Curr Opin Pharmacol. 2024. PMID: 38471384 Review.
Cited by
-
[Generating knowledge from complex data sets in human experimental pain research].Schmerz. 2019 Dec;33(6):502-513. doi: 10.1007/s00482-019-00412-5. Schmerz. 2019. PMID: 31478142 Review. German.
-
Unearthing of Key Genes Driving the Pathogenesis of Alzheimer's Disease via Bioinformatics.Front Genet. 2021 Apr 16;12:641100. doi: 10.3389/fgene.2021.641100. eCollection 2021. Front Genet. 2021. PMID: 33936168 Free PMC article.
-
Genetic study in patients operated dentally and anesthetized with articaine-epinephrine.J Pain Res. 2019 Apr 29;12:1371-1384. doi: 10.2147/JPR.S193745. eCollection 2019. J Pain Res. 2019. PMID: 31118755 Free PMC article.
-
Machine-learned analysis of global and glial/opioid intersection-related DNA methylation in patients with persistent pain after breast cancer surgery.Clin Epigenetics. 2019 Nov 27;11(1):167. doi: 10.1186/s13148-019-0772-4. Clin Epigenetics. 2019. PMID: 31775878 Free PMC article.
-
Artificial intelligence and database for NGS-based diagnosis in rare disease.Front Genet. 2024 Jan 25;14:1258083. doi: 10.3389/fgene.2023.1258083. eCollection 2023. Front Genet. 2024. PMID: 38371307 Free PMC article. Review.
References
-
- Backes C, Harz C, Fischer U, Schmitt J, Ludwig N, Petersen BS, Mueller SC, Kim YJ, Wolf NM, Katus HA, Meder B, Furtwängler R, Franke A, Bohle R, Henn W, Graf N, Keller A, Meese E. New insights into the genetics of glioblastoma multiforme by familial exome sequencing. Oncotarget 2015;6:5918–31. - PMC - PubMed
-
- Bayes M, Price M. An essay towards solving a problem in the doctrine of chances. By the late rev. Mr. Bayes, F. R. S. Communicated by Mr. Price, in a Letter to John Canton, A. M. F. R. S. Philosophical Trans 1763;53:370–418.
-
- Bonferroni CE. Teoria statistica delle classi e calcolo delle probabilita. Pubblicazioni del R Istituto Superiore di Scienze Economiche e Commerciali di Firenze 1936;8:3–62.
-
- Borg I, Groenen P. Modern multidimensional scaling: theory and applications. New York: Springer, 2005.
-
- Boukalova S, Touska F, Marsakova L, Hynkova A, Sura L, Chvojka S, Dittert I, Vlachova V. Gain-of-function mutations in the transient receptor potential channels TRPV1 and TRPA1: how painful? Physiol Res 2014;63(suppl 1):S205–213. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

