Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network
- PMID: 29596349
- PMCID: PMC5979560
- DOI: 10.3390/ijms19041028
Genome-Wide Analyses of the NAC Transcription Factor Gene Family in Pepper (Capsicum annuum L.): Chromosome Location, Phylogeny, Structure, Expression Patterns, Cis-Elements in the Promoter, and Interaction Network
Abstract
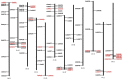
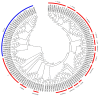
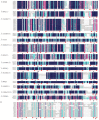
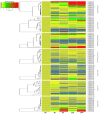
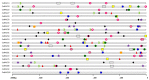
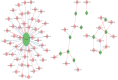
The NAM, ATAF1/2, and CUC2 (NAC) transcription factors form a large plant-specific gene family, which is involved in the regulation of tissue development in response to biotic and abiotic stress. To date, there have been no comprehensive studies investigating chromosomal location, gene structure, gene phylogeny, conserved motifs, or gene expression of NAC in pepper (Capsicum annuum L.). The recent release of the complete genome sequence of pepper allowed us to perform a genome-wide investigation of Capsicum annuum L. NAC (CaNAC) proteins. In the present study, a comprehensive analysis of the CaNAC gene family in pepper was performed, and a total of 104 CaNAC genes were identified. Genome mapping analysis revealed that CaNAC genes were enriched on four chromosomes (chromosomes 1, 2, 3, and 6). In addition, phylogenetic analysis of the NAC domains from pepper, potato, Arabidopsis, and rice showed that CaNAC genes could be clustered into three groups (I, II, and III). Group III, which contained 24 CaNAC genes, was exclusive to the Solanaceae plant family. Gene structure and protein motif analyses showed that these genes were relatively conserved within each subgroup. The number of introns in CaNAC genes varied from 0 to 8, with 83 (78.9%) of CaNAC genes containing two or less introns. Promoter analysis confirmed that CaNAC genes are involved in pepper growth, development, and biotic or abiotic stress responses. Further, the expression of 22 selected CaNAC genes in response to seven different biotic and abiotic stresses [salt, heat shock, drought, Phytophthora capsici, abscisic acid, salicylic acid (SA), and methyl jasmonate (MeJA)] was evaluated by quantitative RT-PCR to determine their stress-related expression patterns. Several putative stress-responsive CaNAC genes, including CaNAC72 and CaNAC27, which are orthologs of the known stress-responsive Arabidopsis gene ANAC055 and potato gene StNAC30, respectively, were highly regulated by treatment with different types of stress. Our results also showed that CaNAC36 plays an important role in the interaction network, interacting with 48 genes. Most of these genes are in the mitogen-activated protein kinase (MAPK) family. Taken together, our results provide a platform for further studies to identify the biological functions of CaNAC genes.
Keywords: NAC family; gene expression; interaction network; pepper; phylogenetic; transcription factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Hisako O., Kouji S., Koji D., Toshifumi N., Yasuhiro O., Kazuo M., Kenichi M., Naoki O., Jun K., Piero C., et al. Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003;10:239–247. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

