Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation
- PMID: 29596410
- PMCID: PMC5892939
- DOI: 10.1371/journal.pbio.2003782
Identifying off-target effects of etomoxir reveals that carnitine palmitoyltransferase I is essential for cancer cell proliferation independent of β-oxidation
Abstract
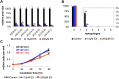
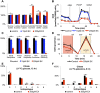
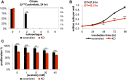
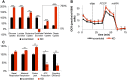
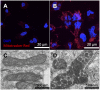
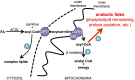
It has been suggested that some cancer cells rely upon fatty acid oxidation (FAO) for energy. Here we show that when FAO was reduced approximately 90% by pharmacological inhibition of carnitine palmitoyltransferase I (CPT1) with low concentrations of etomoxir, the proliferation rate of various cancer cells was unaffected. Efforts to pharmacologically inhibit FAO more than 90% revealed that high concentrations of etomoxir (200 μM) have an off-target effect of inhibiting complex I of the electron transport chain. Surprisingly, however, when FAO was reduced further by genetic knockdown of CPT1, the proliferation rate of these same cells decreased nearly 2-fold and could not be restored by acetate or octanoic acid supplementation. Moreover, CPT1 knockdowns had altered mitochondrial morphology and impaired mitochondrial coupling, whereas cells in which CPT1 had been approximately 90% inhibited by etomoxir did not. Lipidomic profiling of mitochondria isolated from CPT1 knockdowns showed depleted concentrations of complex structural and signaling lipids. Additionally, expression of a catalytically dead CPT1 in CPT1 knockdowns did not restore mitochondrial coupling. Taken together, these results suggest that transport of at least some long-chain fatty acids into the mitochondria by CPT1 may be required for anabolic processes that support healthy mitochondrial function and cancer cell proliferation independent of FAO.
Conflict of interest statement
GJP is a scientific advisory board member for Cambridge Isotope Laboratories. GJP is the recipient of the 2017 Early Career Professor Award from Agilent Technologies. RWG has financial relationships with LipoSpectrum and Platomics.
Figures

References
-
- Schlaepfer IR, Rider L, Rodrigues LU, Gijon MA, Pac CT, Romero L, et al. Lipid catabolism via CPT1 as a therapeutic target for prostate cancer. Mol Cancer Ther. 2014;13(10):2361–71. Epub 2014/08/15. doi: 10.1158/1535-7163.MCT-14-0183 - DOI - PMC - PubMed
-
- Pucci S, Zonetti MJ, Fisco T, Polidoro C, Bocchinfuso G, Palleschi A, et al. Carnitine palmitoyl transferase-1A (CPT1A): a new tumor specific target in human breast cancer. Oncotarget. 2016;7(15):19982–96. Epub 2016/01/23. doi: 10.18632/oncotarget.6964 - DOI - PMC - PubMed
-
- Cirillo A, Di Salle A, Petillo O, Melone MA, Grimaldi G, Bellotti A, et al. High grade glioblastoma is associated with aberrant expression of ZFP57, a protein involved in gene imprinting, and of CPT1A and CPT1C that regulate fatty acid metabolism. Cancer Biol Ther. 2014;15(6):735–41. Epub 2014/03/13. doi: 10.4161/cbt.28408 - DOI - PMC - PubMed
-
- Zaugg K, Yao Y, Reilly PT, Kannan K, Kiarash R, Mason J, et al. Carnitine palmitoyltransferase 1C promotes cell survival and tumor growth under conditions of metabolic stress. Genes Dev. 2011;25(10):1041–51. Epub 2011/05/18. doi: 10.1101/gad.1987211 - DOI - PMC - PubMed
-
- Tirado-Velez JM, Joumady I, Saez-Benito A, Cozar-Castellano I, Perdomo G. Inhibition of fatty acid metabolism reduces human myeloma cells proliferation. PloS one. 2012;7(9):e46484 Epub 2012/10/03. doi: 10.1371/journal.pone.0046484 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

