Pluripotency gene network dynamics: System views from parametric analysis
- PMID: 29596533
- PMCID: PMC5875786
- DOI: 10.1371/journal.pone.0194464
Pluripotency gene network dynamics: System views from parametric analysis
Abstract
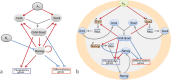
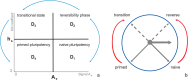
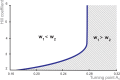
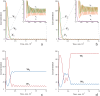
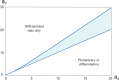
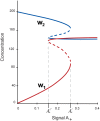
Multiple experimental data demonstrated that the core gene network orchestrating self-renewal and differentiation of mouse embryonic stem cells involves activity of Oct4, Sox2 and Nanog genes by means of a number of positive feedback loops among them. However, recent studies indicated that the architecture of the core gene network should also incorporate negative Nanog autoregulation and might not include positive feedbacks from Nanog to Oct4 and Sox2. Thorough parametric analysis of the mathematical model based on this revisited core regulatory circuit identified that there are substantial changes in model dynamics occurred depending on the strength of Oct4 and Sox2 activation and molecular complexity of Nanog autorepression. The analysis showed the existence of four dynamical domains with different numbers of stable and unstable steady states. We hypothesize that these domains can constitute the checkpoints in a developmental progression from naïve to primed pluripotency and vice versa. During this transition, parametric conditions exist, which generate an oscillatory behavior of the system explaining heterogeneity in expression of pluripotent and differentiation factors in serum ESC cultures. Eventually, simulations showed that addition of positive feedbacks from Nanog to Oct4 and Sox2 leads mainly to increase of the parametric space for the naïve ESC state, in which pluripotency factors are strongly expressed while differentiation ones are repressed.
Conflict of interest statement
Figures


References
-
- Smith A. (2017). Formative pluripotency: the executive phase in a developmental continuum. Development, 144(3), 365–373. doi: 10.1242/dev.142679 - DOI - PMC - PubMed
-
- Li M., Belmonte J. C. I. (2017). Ground rules of the pluripotency gene regulatory network. Nature Reviews Genetics. - PubMed
-
- Betschinger J. (2016). Charting developmental dissolution of pluripotency. Journal of Molecular Biology. - PubMed
-
- Makarev E., Fortney K., Litovchenko M., Braunewell K. H., Zhavoronkov A., Atala A. (2015). Quantifying signaling pathway activation to monitor the quality of induced pluripotent stem cells. Oncotarget, 6(27), 23204 doi: 10.18632/oncotarget.4673 - DOI - PMC - PubMed
-
- van den Hurk M., Kenis G., Bardy C., van den Hove D. L., Gage F. H., Steinbusch H. W., et al. (2016). Transcriptional and epigenetic mechanisms of cellular reprogramming to induced pluripotency. Epigenomics, 8(8), 1131–1149. doi: 10.2217/epi-2016-0032 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

