The DNA damage response activates HPV16 late gene expression at the level of RNA processing
- PMID: 29596642
- PMCID: PMC6007495
- DOI: 10.1093/nar/gky227
The DNA damage response activates HPV16 late gene expression at the level of RNA processing
Abstract
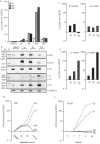
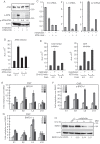
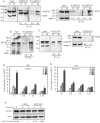
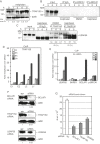
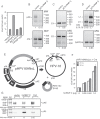
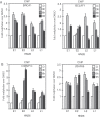
We show that the alkylating cancer drug melphalan activated the DNA damage response and induced human papillomavirus type 16 (HPV16) late gene expression in an ATM- and Chk1/2-dependent manner. Activation of HPV16 late gene expression included inhibition of the HPV16 early polyadenylation signal that resulted in read-through into the late region of HPV16. This was followed by activation of the exclusively late, HPV16 splice sites SD3632 and SA5639 and production of spliced late L1 mRNAs. Altered HPV16 mRNA processing was paralleled by increased association of phosphorylated BRCA1, BARD1, BCLAF1 and TRAP150 with HPV16 DNA, and increased association of RNA processing factors U2AF65 and hnRNP C with HPV16 mRNAs. These RNA processing factors inhibited HPV16 early polyadenylation and enhanced HPV16 late mRNA splicing, thereby activating HPV16 late gene expression.
Figures





References
-
- Chow L.T., Broker T.R., Steinberg B.M.. The natural history of human papillomavirus infections of the mucosal epithelia. APMIS. 2010; 118:422–449. - PubMed
-
- Schiffman M., Doorbar J., Wentzensen N., de Sanjose S., Fakhry C., Monk B.J., Stanley M.A., Franceschi S.. Carcinogenic human papillomavirus infection. Nat. Rev. Dis. Primers. 2016; 2:16086. - PubMed
-
- Bouvard V., Baan R., Straif K., Grosse Y., Secretan B., El Ghissassi F., Benbrahim-Tallaa L., Guha N., Freeman C., Galichet L. et al. A review of human carcinogens–Part B: biological agents. Lancet Oncol. 2009; 10:321–322. - PubMed
-
- Walboomers J.M., Jacobs M.V., Manos M.M., Bosch F.X., Kummer J.A., Shah K.V., Snijders P.J., Peto J., Meijer C.J., Munoz N.. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999; 189:12–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

