Signal peptides for recombinant protein secretion in bacterial expression systems
- PMID: 29598818
- PMCID: PMC5875014
- DOI: 10.1186/s12934-018-0901-3
Signal peptides for recombinant protein secretion in bacterial expression systems
Abstract
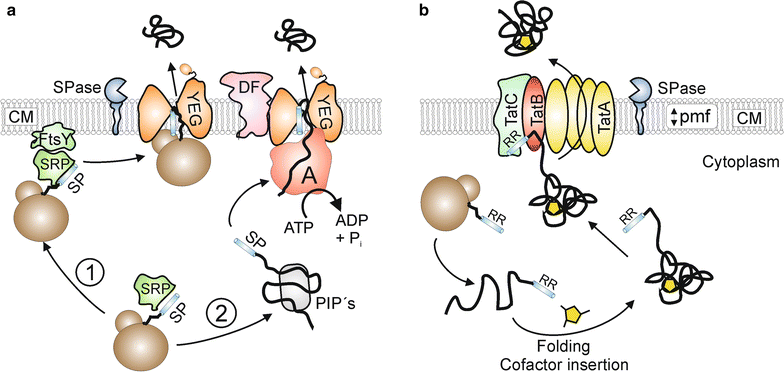
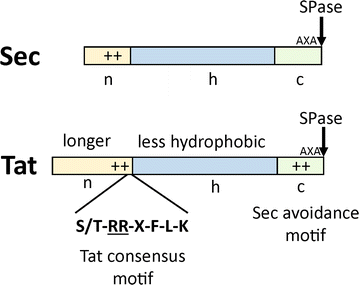
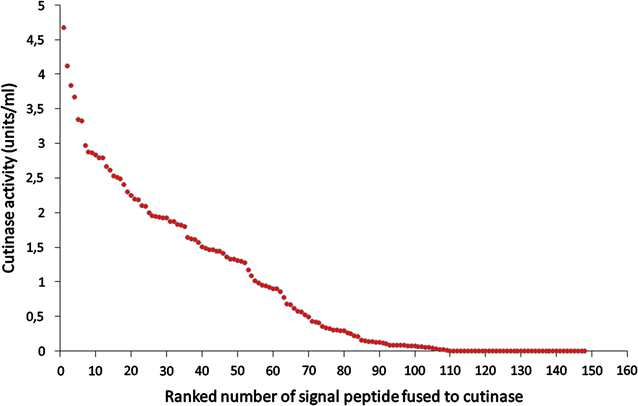
The secretion of biotechnologically or pharmaceutically relevant recombinant proteins into the culture supernatant of a bacterial expression host greatly facilitates their downstream processing and significantly reduces the production costs. The first step during the secretion of a desired target protein into the growth medium is its transport across the cytoplasmic membrane. In bacteria, two major export pathways, the general secretion or Sec pathway and the twin-arginine translocation or Tat pathway, exist for the transport of proteins across the plasma membrane. The routing into one of these alternative protein export systems requires the fusion of a Sec- or Tat-specific signal peptide to the amino-terminal end of the desired target protein. Since signal peptides, besides being required for the targeting to and membrane translocation by the respective protein translocases, also have additional influences on the biosynthesis, the folding kinetics, and the stability of the respective target proteins, it is not possible so far to predict in advance which signal peptide will perform best in the context of a given target protein and a given bacterial expression host. As outlined in this review, the most promising way to find the optimal signal peptide for a desired protein is to screen the largest possible diversity of signal peptides, either generated by signal peptide variation using large signal peptide libraries or, alternatively, by optimization of a given signal peptide using site-directed or random mutagenesis strategies.
Keywords: Gram-positive bacteria; Protein secretion; Recombinant protein production; Sec pathway; Signal peptide; Twin-arginine-translocation (Tat) pathway.
Figures
Similar articles
-
Signal Peptide Hydrophobicity Modulates Interaction with the Twin-Arginine Translocase.mBio. 2017 Aug 1;8(4):e00909-17. doi: 10.1128/mBio.00909-17. mBio. 2017. PMID: 28765221 Free PMC article.
-
Comparative analysis of twin-arginine (Tat)-dependent protein secretion of a heterologous model protein (GFP) in three different Gram-positive bacteria.Appl Microbiol Biotechnol. 2007 Sep;76(3):633-42. doi: 10.1007/s00253-007-0934-8. Epub 2007 Apr 24. Appl Microbiol Biotechnol. 2007. PMID: 17453196
-
Beyond amino acids: Use of the Corynebacterium glutamicum cell factory for the secretion of heterologous proteins.J Biotechnol. 2017 Sep 20;258:101-109. doi: 10.1016/j.jbiotec.2017.02.023. Epub 2017 Feb 24. J Biotechnol. 2017. PMID: 28238807
-
Leaving home ain't easy: protein export systems in Gram-positive bacteria.Res Microbiol. 2013 Jul-Aug;164(6):664-74. doi: 10.1016/j.resmic.2013.03.014. Epub 2013 Mar 26. Res Microbiol. 2013. PMID: 23541477 Review.
-
Functional genomic analysis of the Bacillus subtilis Tat pathway for protein secretion.J Biotechnol. 2002 Sep 25;98(2-3):243-54. doi: 10.1016/s0168-1656(02)00135-9. J Biotechnol. 2002. PMID: 12141990 Review.
Cited by
-
Light inducible gene expression system for Streptomyces.Sci Rep. 2024 Oct 28;14(1):25852. doi: 10.1038/s41598-024-76860-6. Sci Rep. 2024. PMID: 39468183 Free PMC article.
-
TISIGNER.com: web services for improving recombinant protein production.Nucleic Acids Res. 2021 Jul 2;49(W1):W654-W661. doi: 10.1093/nar/gkab175. Nucleic Acids Res. 2021. PMID: 33744969 Free PMC article.
-
Novel Enhanced Mammalian Cell Transient Expression Vector via Promoter Combination.Int J Mol Sci. 2024 Feb 16;25(4):2330. doi: 10.3390/ijms25042330. Int J Mol Sci. 2024. PMID: 38397006 Free PMC article.
-
Tuning recombinant protein expression to match secretion capacity.Microb Cell Fact. 2018 Dec 22;17(1):199. doi: 10.1186/s12934-018-1047-z. Microb Cell Fact. 2018. PMID: 30577801 Free PMC article.
-
Membrane vesicle engineering with "à la carte" bacterial-immunogenic molecules for organism-free plant vaccination.Microb Biotechnol. 2023 Dec;16(12):2223-2235. doi: 10.1111/1751-7915.14323. Epub 2023 Aug 2. Microb Biotechnol. 2023. PMID: 37530752 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

