Combined flow cytometry and high-throughput image analysis for the study of essential genes in Caenorhabditis elegans
- PMID: 29598825
- PMCID: PMC5875015
- DOI: 10.1186/s12915-018-0496-5
Combined flow cytometry and high-throughput image analysis for the study of essential genes in Caenorhabditis elegans
Abstract
Background: Advances in automated image-based microscopy platforms coupled with high-throughput liquid workflows have facilitated the design of large-scale screens utilising multicellular model organisms such as Caenorhabditis elegans to identify genetic interactions, therapeutic drugs or disease modifiers. However, the analysis of essential genes has lagged behind because lethal or sterile mutations pose a bottleneck for high-throughput approaches, and a systematic way to analyse genetic interactions of essential genes in multicellular organisms has been lacking.
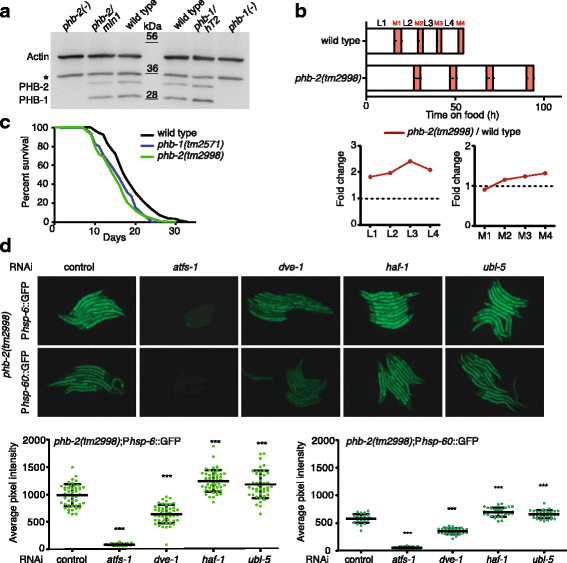
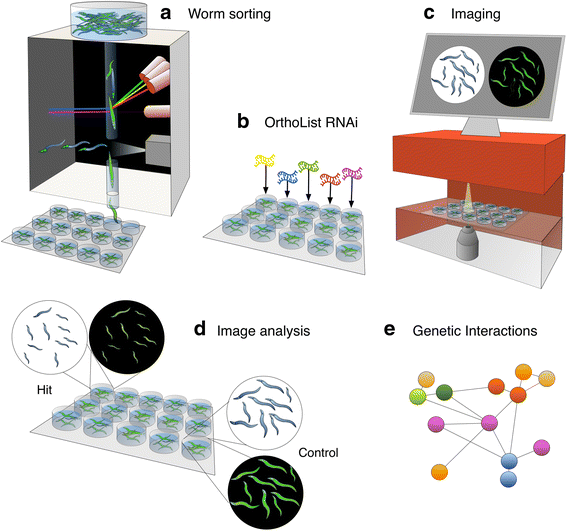
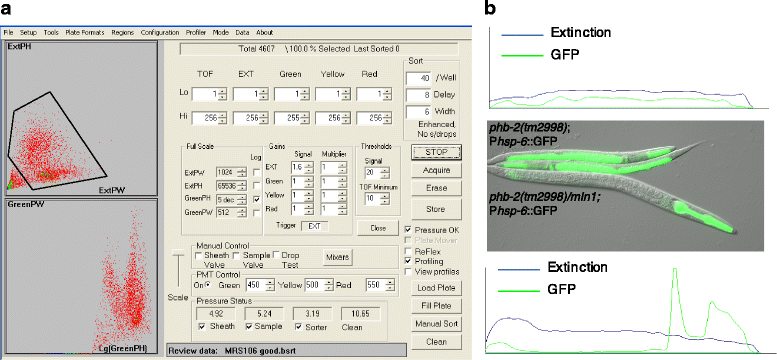
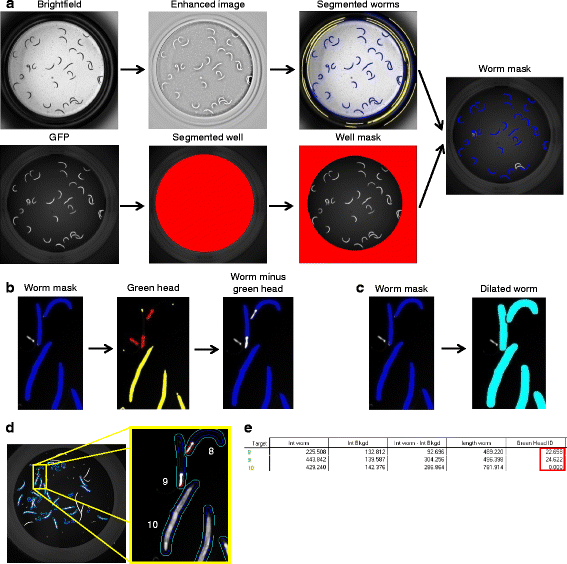
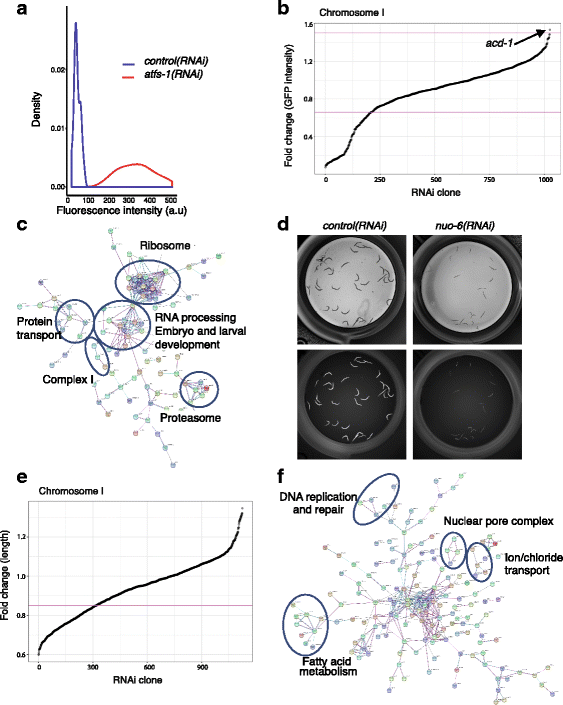
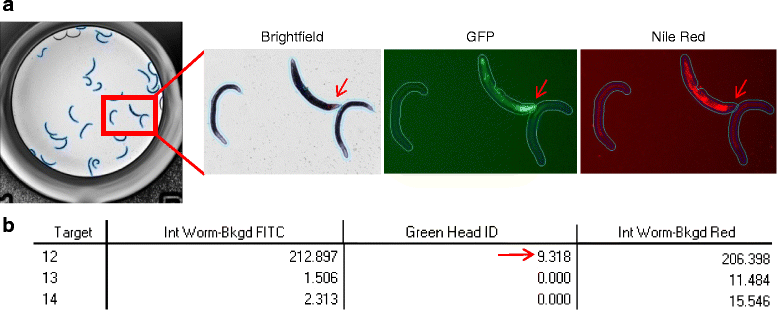
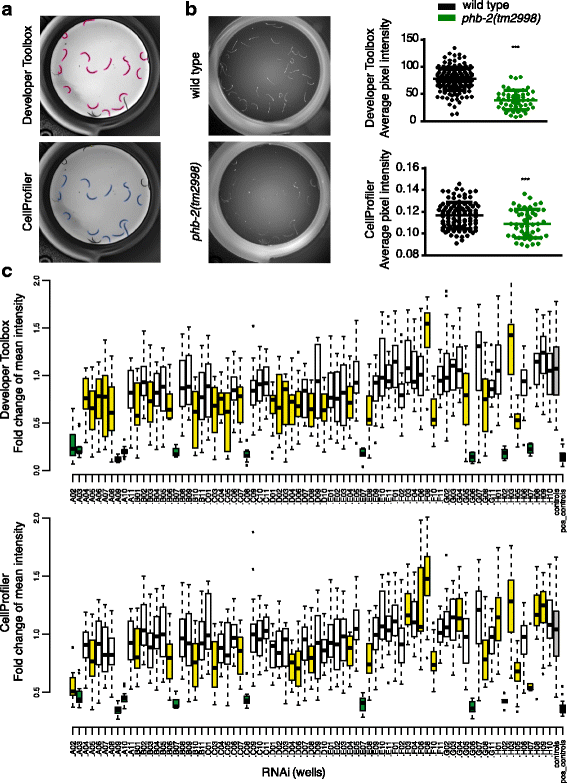
Results: In C. elegans, non-conditional lethal mutations can be maintained in heterozygosity using chromosome balancers, commonly expressing green fluorescent protein (GFP) in the pharynx. However, gene expression or function is typically monitored by the use of fluorescent reporters marked with the same fluorophore, presenting a challenge to sort worm populations of interest, particularly at early larval stages. Here, we develop a sorting strategy capable of selecting homozygous mutants carrying a GFP stress reporter from GFP-balanced animals at the second larval stage. Because sorting is not completely error-free, we develop an automated high-throughput image analysis protocol that identifies and discards animals carrying the chromosome balancer. We demonstrate the experimental usefulness of combining sorting of homozygous lethal mutants and automated image analysis in a functional genomic RNA interference (RNAi) screen for genes that genetically interact with mitochondrial prohibitin (PHB). Lack of PHB results in embryonic lethality, while homozygous PHB deletion mutants develop into sterile adults due to maternal contribution and strongly induce the mitochondrial unfolded protein response (UPRmt). In a chromosome-wide RNAi screen for C. elegans genes having human orthologues, we uncover both known and new PHB genetic interactors affecting the UPRmt and growth.
Conclusions: The method presented here allows the study of balanced lethal mutations in a high-throughput manner. It can be easily adapted depending on the user's requirements and should serve as a useful resource for the C. elegans community for probing new biological aspects of essential nematode genes as well as the generation of more comprehensive genetic networks.
Keywords: C. elegans; Essential genes; High-content; High-throughput; Image analysis; Mitochondria; Prohibitins; Screens; UPRmt; Worm sorting.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Methods to Study the Mitochondrial Unfolded Protein Response (UPRmt) in Caenorhabditis elegans.Methods Mol Biol. 2022;2378:249-259. doi: 10.1007/978-1-0716-1732-8_16. Methods Mol Biol. 2022. PMID: 34985705
-
"High-Throughput Characterization of Region-Specific Mitochondrial Function and Morphology".Sci Rep. 2017 Jul 27;7(1):6749. doi: 10.1038/s41598-017-05152-z. Sci Rep. 2017. PMID: 28751733 Free PMC article.
-
Activation of the mitochondrial unfolded protein response does not predict longevity in Caenorhabditis elegans.Nat Commun. 2014 Mar 24;5:3483. doi: 10.1038/ncomms4483. Nat Commun. 2014. PMID: 24662282 Free PMC article.
-
Large-scale gene expression pattern analysis, in situ, in Caenorhabditis elegans.Brief Funct Genomic Proteomic. 2008 May;7(3):175-83. doi: 10.1093/bfgp/eln013. Epub 2008 Mar 9. Brief Funct Genomic Proteomic. 2008. PMID: 18332038 Review.
-
Functional genomic approaches using the nematode Caenorhabditis elegans as a model system.J Biochem Mol Biol. 2004 Jan 31;37(1):107-13. doi: 10.5483/bmbrep.2004.37.1.107. J Biochem Mol Biol. 2004. PMID: 14761308 Review.
Cited by
-
Microfluidic-Assisted Caenorhabditis elegans Sorting: Current Status and Future Prospects.Cyborg Bionic Syst. 2023 Apr 14;4:0011. doi: 10.34133/cbsystems.0011. eCollection 2023. Cyborg Bionic Syst. 2023. PMID: 37287459 Free PMC article.
-
OrthoList 2: A New Comparative Genomic Analysis of Human and Caenorhabditis elegans Genes.Genetics. 2018 Oct;210(2):445-461. doi: 10.1534/genetics.118.301307. Epub 2018 Aug 17. Genetics. 2018. PMID: 30120140 Free PMC article.
-
Mitochondrial Quality Control Mechanisms and the PHB (Prohibitin) Complex.Cells. 2018 Nov 29;7(12):238. doi: 10.3390/cells7120238. Cells. 2018. PMID: 30501123 Free PMC article. Review.
-
Fluorizoline-induced apoptosis requires prohibitins in nematodes and human cells.Apoptosis. 2021 Feb;26(1-2):83-95. doi: 10.1007/s10495-020-01651-z. Epub 2021 Jan 2. Apoptosis. 2021. PMID: 33387147
-
Artificial intelligence in imaging flow cytometry.Front Bioinform. 2023 Oct 9;3:1229052. doi: 10.3389/fbinf.2023.1229052. eCollection 2023. Front Bioinform. 2023. PMID: 37877042 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

