A Structured Workflow for Mapping Human Sin3 Histone Deacetylase Complex Interactions Using Halo-MudPIT Affinity-Purification Mass Spectrometry
- PMID: 29599190
- PMCID: PMC6030732
- DOI: 10.1074/mcp.TIR118.000661
A Structured Workflow for Mapping Human Sin3 Histone Deacetylase Complex Interactions Using Halo-MudPIT Affinity-Purification Mass Spectrometry
Abstract
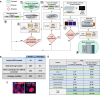
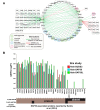
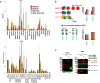
Although a variety of affinity purification mass spectrometry (AP-MS) strategies have been used to investigate complex interactions, many of these are susceptible to artifacts because of substantial overexpression of the exogenously expressed bait protein. Here we present a logical and systematic workflow that uses the multifunctional Halo tag to assess the correct localization and behavior of tagged subunits of the Sin3 histone deacetylase complex prior to further AP-MS analysis. Using this workflow, we modified our tagging/expression strategy with 21.7% of the tagged bait proteins that we constructed, allowing us to quickly develop validated reagents. Specifically, we apply the workflow to map interactions between stably expressed versions of the Sin3 subunits SUDS3, SAP30, or SAP30L and other cellular proteins. Here we show that the SAP30 and SAP30L paralogues strongly associate with the core Sin3 complex, but SAP30L has unique associations with the proteasome and the myelin sheath. Next, we demonstrate an advancement of the complex NSAF (cNSAF) approach, in which normalization to the scaffold protein SIN3A accounts for variations in the proportion of each bait capturing Sin3 complexes and allows a comparison among different baits capturing the same protein complex. This analysis reveals that although the Sin3 subunit SUDS3 appears to be used in both SIN3A and SIN3B based complexes, the SAP30 subunit is not used in SIN3B based complexes. Intriguingly, we do not detect the Sin3 subunits SAP18 and SAP25 among the 128 high-confidence interactions identified, suggesting that these subunits may not be common to all versions of the Sin3 complex in human cells. This workflow provides the framework for building validated reagents to assemble quantitative interaction networks for chromatin remodeling complexes and provides novel insights into focused protein interaction networks.
Keywords: Affinity tagging; Chromatin function or biology; Imaging; Label-free quantification; Macromolecular complex analysis; Networks*; Protein complex analysis; Protein-Protein Interactions*.
© 2018 Banks et al.
Figures



References
-
- Hein M. Y., Hubner N. C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I. A., Weisswange I., Mansfeld J., Buchholz F., Hyman A. A., and Mann M. (2015) A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell 163, 712–723 - PubMed
-
- Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., Erickson B. K., Wühr M., Chick J., Zhai B., Kolippakkam D., Mintseris J., Obar R. A., Harris T., Artavanis-Tsakonas S., Sowa M. E., De Camilli P., Paulo J. A., Harper J. W., and Gygi S. P. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 - PMC - PubMed
-
- Sardiu M. E., Smith K. T., Groppe B. D., Gilmore J. M., Saraf A., Egidy R., Peak A., Seidel C. W., Florens L., Workman J. L., and Washburn M. P. (2014) Suberoylanilide hydroxamic acid (SAHA)-induced dynamics of a human histone deacetylase protein interaction network. Mol. Cell. Proteomics 13, 3114–3125 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

