TagF-mediated repression of bacterial type VI secretion systems involves a direct interaction with the cytoplasmic protein Fha
- PMID: 29599293
- PMCID: PMC5995506
- DOI: 10.1074/jbc.RA117.001618
TagF-mediated repression of bacterial type VI secretion systems involves a direct interaction with the cytoplasmic protein Fha
Abstract
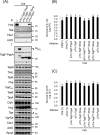
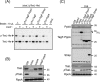
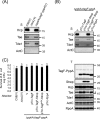
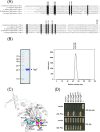
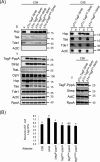
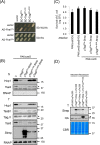
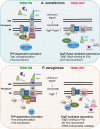
The bacterial type VI secretion system (T6SS) delivers effectors into eukaryotic host cells or toxins into bacterial competitor for survival and fitness. The T6SS is positively regulated by the threonine phosphorylation pathway (TPP) and negatively by the T6SS-accessory protein TagF. Here, we studied the mechanisms underlying TagF-mediated T6SS repression in two distinct bacterial pathogens, Agrobacterium tumefaciens and Pseudomonas aeruginosa. We found that in A. tumefaciens, T6SS toxin secretion and T6SS-dependent antibacterial activity are suppressed by a two-domain chimeric protein consisting of TagF and PppA, a putative phosphatase. Remarkably, this TagF domain is sufficient to post-translationally repress the T6SS, and this inhibition is independent of TPP. This repression requires interaction with a cytoplasmic protein, Fha, critical for activating T6SS assembly. In P. aeruginosa, PppA and TagF are two distinct proteins that repress T6SS in TPP-dependent and -independent pathways, respectively. P. aeruginosa TagF interacts with Fha1, suggesting that formation of this complex represents a conserved TagF-mediated regulatory mechanism. Using TagF variants with substitutions of conserved amino acid residues at predicted protein-protein interaction interfaces, we uncovered evidence that the TagF-Fha interaction is critical for TagF-mediated T6SS repression in both bacteria. TagF inhibits T6SS without affecting T6SS protein abundance in A. tumefaciens, but TagF overexpression reduces the protein levels of all analyzed T6SS components in P. aeruginosa Our results indicate that TagF interacts with Fha, which in turn could impact different stages of T6SS assembly in different bacteria, possibly reflecting an evolutionary divergence in T6SS control.
Keywords: Agrobacterium tumefaciens; Fha; PppA; Pseudomonas aeruginosa; TagF; antibacterial activity; bacterial genetics; gene regulation; post-translational regulation; protein phosphorylation; protein secretion; protein-protein interaction; type VI secretion system.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
Killing with proficiency: Integrated post-translational regulation of an offensive Type VI secretion system.PLoS Pathog. 2018 Jul 27;14(7):e1007230. doi: 10.1371/journal.ppat.1007230. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30052683 Free PMC article.
-
Crystal structure of PppA from Pseudomonas aeruginosa, a key regulatory component of type VI secretion systems.Biochem Biophys Res Commun. 2019 Aug 13;516(1):196-201. doi: 10.1016/j.bbrc.2019.06.020. Epub 2019 Jun 14. Biochem Biophys Res Commun. 2019. PMID: 31208722
-
Fha interaction with phosphothreonine of TssL activates type VI secretion in Agrobacterium tumefaciens.PLoS Pathog. 2014 Mar 13;10(3):e1003991. doi: 10.1371/journal.ppat.1003991. eCollection 2014 Mar. PLoS Pathog. 2014. PMID: 24626341 Free PMC article.
-
Composition, function, and regulation of T6SS in Pseudomonas aeruginosa.Microbiol Res. 2015 Mar;172:19-25. doi: 10.1016/j.micres.2015.01.004. Epub 2015 Jan 7. Microbiol Res. 2015. PMID: 25721475 Review.
-
The Agrobacterium Type VI Secretion System: A Contractile Nanomachine for Interbacterial Competition.Curr Top Microbiol Immunol. 2018;418:215-231. doi: 10.1007/82_2018_99. Curr Top Microbiol Immunol. 2018. PMID: 29992360 Review.
Cited by
-
EspM Is a Conserved Transcription Factor That Regulates Gene Expression in Response to the ESX-1 System.mBio. 2020 Feb 4;11(1):e02807-19. doi: 10.1128/mBio.02807-19. mBio. 2020. PMID: 32019792 Free PMC article.
-
The Bradyrhizobium Sp. LmicA16 Type VI Secretion System Is Required for Efficient Nodulation of Lupinus Spp.Microb Ecol. 2022 Oct;84(3):844-855. doi: 10.1007/s00248-021-01892-8. Epub 2021 Oct 25. Microb Ecol. 2022. PMID: 34697646
-
Functional Analysis of EspM, an ESX-1-Associated Transcription Factor in Mycobacterium marinum.J Bacteriol. 2022 Dec 20;204(12):e0023322. doi: 10.1128/jb.00233-22. Epub 2022 Nov 30. J Bacteriol. 2022. PMID: 36448785 Free PMC article.
-
Killing with proficiency: Integrated post-translational regulation of an offensive Type VI secretion system.PLoS Pathog. 2018 Jul 27;14(7):e1007230. doi: 10.1371/journal.ppat.1007230. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 30052683 Free PMC article.
-
T6SS nuclease effectors in Pseudomonas syringae act as potent antimicrobials in interbacterial competition.J Bacteriol. 2024 Jun 20;206(6):e0027323. doi: 10.1128/jb.00273-23. Epub 2024 May 8. J Bacteriol. 2024. PMID: 38717111 Free PMC article.
References
-
- Ma L. S., Lin J. S., and Lai E. M. (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191, 4316–4329 10.1128/JB.00029-09 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

