Improved efficiency of in situ protein analysis by proximity ligation using UnFold probes
- PMID: 29599435
- PMCID: PMC5876389
- DOI: 10.1038/s41598-018-23582-1
Improved efficiency of in situ protein analysis by proximity ligation using UnFold probes
Abstract
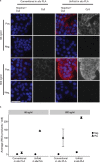
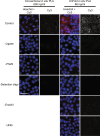
We have redesigned probes for in situ proximity ligation assay (PLA), resulting in more efficient localized detection of target proteins. In situ PLA depends on recognition of target proteins by pairs of antibody-oligonucleotide conjugates (PLA probes), which jointly give rise to DNA circles that template localized rolling circle amplification reactions. The requirement for dual recognition of the target proteins improves selectivity by ignoring any cross-reactivity not shared by the antibodies, and it allows detection of protein-protein interactions and post-translational modifications. We herein describe an improved design of the PLA probes -UnFold probes - where all elements required for formation of circular DNA strands are incorporated in the probes. Premature interactions between the UnFold probes are prevented by including an enzymatic "unfolding" step in the detection reactions. This allows DNA circles to form by pairs of reagents only after excess reagents have been removed. We demonstrate the performance of UnFold probes for detection of protein-protein interactions and post-translational modifications in fixed cells and tissues, revealing considerably more efficient signal generation. We also apply the UnFold probes to detect IL-6 in solution phase after capture on solid supports, demonstrating increased sensitivity over both normal sandwich enzyme-linked immunosorbent assays and conventional PLA assays.
Conflict of interest statement
U.L. is a founder and shareholder of Olink Biosciences having rights to the PLA technology. O.S. and U.L. are inventors of a patent covering the UnFold probe invention. A.K., K.G., T.E., J.H., B.K., M.L., D.R., J.O. and L.A. declare no competing interests.
Figures





References
-
- Coons A, Creech H, Jones RN, Berliner E. Demonstration of pneumoccocal antigen in tissues by use of fluorescent antibody. J Immunol. 1942;45:12.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

