The successes and future prospects of the linear antisense RNA amplification methodology
- PMID: 29599441
- PMCID: PMC7086549
- DOI: 10.1038/nprot.2018.011
The successes and future prospects of the linear antisense RNA amplification methodology
Abstract
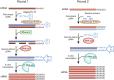
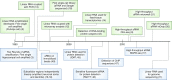
It has been over a quarter of a century since the introduction of the linear RNA amplification methodology known as antisense RNA (aRNA) amplification. Whereas most molecular biology techniques are rapidly replaced owing to the fast-moving nature of development in the field, the aRNA procedure has become a base that can be built upon through varied uses of the technology. The technique was originally developed to assess RNA populations from small amounts of starting material, including single cells, but over time its use has evolved to include the detection of various cellular entities such as proteins, RNA-binding-protein-associated cargoes, and genomic DNA. In this Perspective we detail the linear aRNA amplification procedure and its use in assessing various components of a cell's chemical phenotype. This procedure is particularly useful in efforts to multiplex the simultaneous detection of various cellular processes. These efforts are necessary to identify the quantitative chemical phenotype of cells that underlies cellular function.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

Similar articles
-
aRNA-LongSAGE: SAGE with antisense RNA.Methods Mol Biol. 2008;387:39-54. doi: 10.1007/978-1-59745-454-4_3. Methods Mol Biol. 2008. PMID: 18287621
-
Antisense RNA amplification for target assessment of total mRNA from a single cell.Cold Spring Harb Protoc. 2014 Nov 3;2014(11):1149-60. doi: 10.1101/pdb.prot072454. Cold Spring Harb Protoc. 2014. PMID: 25368303
-
Manual microdissection combined with antisense RNA-longSAGE for the analysis of limited cell numbers.Methods Mol Biol. 2010;576:135-54. doi: 10.1007/978-1-59745-545-9_8. Methods Mol Biol. 2010. PMID: 19882261
-
Expression profiling using human tissues in combination with RNA amplification and microarray analysis: assessment of Langerhans cell histiocytosis.Amino Acids. 2005 May;28(3):279-90. doi: 10.1007/s00726-005-0177-x. Epub 2005 Mar 30. Amino Acids. 2005. PMID: 15791395 Review.
-
Strategies for microarray analysis of limiting amounts of RNA.Brief Funct Genomic Proteomic. 2003 Apr;2(1):31-6. doi: 10.1093/bfgp/2.1.31. Brief Funct Genomic Proteomic. 2003. PMID: 15239941 Review.
Cited by
-
LINCATRA: Two-cycle method to amplify RNA for transcriptome analysis from formalin-fixed paraffin-embedded tissue.Heliyon. 2024 Jun 12;10(12):e32896. doi: 10.1016/j.heliyon.2024.e32896. eCollection 2024 Jun 30. Heliyon. 2024. PMID: 38988576 Free PMC article.
-
Subcellular omics: a new frontier pushing the limits of resolution, complexity and throughput.Nat Methods. 2023 Mar;20(3):331-335. doi: 10.1038/s41592-023-01788-0. Nat Methods. 2023. PMID: 36899160 Free PMC article.
-
Leading the pack: Best practices in comparative canine cancer genomics to inform human oncology.Vet Comp Oncol. 2023 Dec;21(4):565-577. doi: 10.1111/vco.12935. Epub 2023 Oct 1. Vet Comp Oncol. 2023. PMID: 37778398 Free PMC article. Review.
-
Deep scRNA sequencing reveals a broadly applicable Regeneration Classifier and implicates antioxidant response in corticospinal axon regeneration.Neuron. 2023 Dec 20;111(24):3953-3969.e5. doi: 10.1016/j.neuron.2023.09.019. Epub 2023 Oct 16. Neuron. 2023. PMID: 37848024 Free PMC article.
-
Future of Digital Assays to Resolve Clinical Heterogeneity of Single Extracellular Vesicles.ACS Nano. 2022 Aug 23;16(8):11619-11645. doi: 10.1021/acsnano.2c04337. Epub 2022 Jul 29. ACS Nano. 2022. PMID: 35904433 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

