The emerging clinical relevance of genomics in cancer medicine
- PMID: 29599476
- PMCID: PMC6658089
- DOI: 10.1038/s41571-018-0002-6
The emerging clinical relevance of genomics in cancer medicine
Abstract
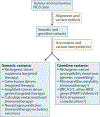
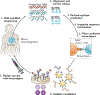
The combination of next-generation sequencing and advanced computational data analysis approaches has revolutionized our understanding of the genomic underpinnings of cancer development and progression. The coincident development of targeted small molecule and antibody-based therapies that target a cancer's genomic dependencies has fuelled the transition of genomic assays into clinical use in patients with cancer. Beyond the identification of individual targetable alterations, genomic methods can gauge mutational load, which might predict a therapeutic response to immune-checkpoint inhibitors or identify cancer-specific proteins that inform the design of personalized anticancer vaccines. Emerging clinical applications of cancer genomics include monitoring treatment responses and characterizing mechanisms of resistance. The increasing relevance of genomics to clinical cancer care also highlights several considerable challenges, including the need to promote equal access to genomic testing.
Figures

References
-
- Garraway LA & Lander ES Lessons from the cancer genome. Cell 153, 17–37 (2013). - PubMed
-
- Meyerson M, Gabriel S & Getz G Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 11, 685–696 (2010). - PubMed
-
- Do H & Dobrovic A Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin. Chem. 61, 64–71 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

