Enterobacter bugandensis: a novel enterobacterial species associated with severe clinical infection
- PMID: 29599516
- PMCID: PMC5876403
- DOI: 10.1038/s41598-018-23069-z
Enterobacter bugandensis: a novel enterobacterial species associated with severe clinical infection
Abstract
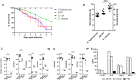
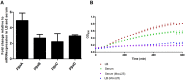
Nosocomial pathogens can cause life-threatening infections in neonates and immunocompromised patients. E. bugandensis (EB-247) is a recently described species of Enterobacter, associated with neonatal sepsis. Here we demonstrate that the extended spectrum ß-lactam (ESBL) producing isolate EB-247 is highly virulent in both Galleria mellonella and mouse models of infection. Infection studies in a streptomycin-treated mouse model showed that EB-247 is as efficient as Salmonella Typhimurium in inducing systemic infection and release of proinflammatory cytokines. Sequencing and analysis of the complete genome and plasmid revealed that virulence properties are associated with the chromosome, while antibiotic-resistance genes are exclusively present on a 299 kb IncHI plasmid. EB-247 grew in high concentrations of human serum indicating septicemic potential. Using whole genome-based transcriptome analysis we found 7% of the genome was mobilized for growth in serum. Upregulated genes include those involved in the iron uptake and storage as well as metabolism. The lasso peptide microcin J25 (MccJ25), an inhibitor of iron-uptake and RNA polymerase activity, inhibited EB-247 growth. Our studies indicate that Enterobacter bugandensis is a highly pathogenic species of the genus Enterobacter. Further studies on the colonization and virulence potential of E. bugandensis and its association with septicemic infection is now warranted.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

